Research
Challenges in Cancer Biology
Cancer is the second leading cause of death in the United States. Malignant tumors are difficult to treat due to drug resistance and their ability to undergo metastasis. These challenges demand studies on cancer as a dynamic process, with an increased focus on understanding tumor initiation and progression mechanisms. Information from such studies will allow the detection of pre-cancerous lesions to move the first line of defense prior to full-blown malignancy.
A few key questions to be answered:
- Where and when does a tumor initiate?
- What are the unique molecular and cellular features of tumor-initiating cells?
- How do tumor cells interact and co-evolve with their environment?
- How do tumor cells migrate to metastasize?
- How can we use the information to detect, prevent, and treat cancer?
The importance of using mouse models to study cancers
Though cancers can be studied with many approaches, the answers to questions above require us to perform studies in native settings of cancers. Because one cannot study human patients unless they carry detectable tumors, it becomes very important to investigate these questions using mouse cancer models. One important event for human tumor initiation is the loss of heterozygosity (LOH) of tumor suppressor genes, which has been modeled in mice using gene knockout technique. However, current mouse genetic models have difficulties in controlling gene inactivation in a small number of cells to mimic the sporadic nature of the LOH events typical of human tumors. Even if gene inactivation can be limited to a few cells, it is very difficult to unambiguously distinguish them from surrounding cells for mechanistic analysis. Therefore, it becomes very important for cancer researchers to develop more physiologically relevant mouse models to address the critical questions raised above.
Use mouse genetic mosaic system to model cancers
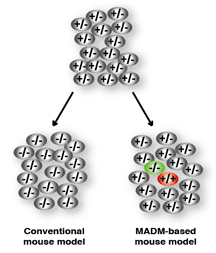
The research theme in the Zong lab is to use a mouse genetic mosaic system termed MADM (Mosaic Analysis with Double Markers) [Zong 2005 Cell] to probe into core cancer mechanisms. Rather than generating more TSG KO mouse models, MADM pushes the phenotypic analysis of currently available models to an unprecedented resolution [Muzumdar 2007 PNAS]. After recombining an available TSG KO allele onto a MADM-bearing chromosome, the MADM system generates sporadic mutant cells from a heterozygous mouse and uniquely labels the mutant cells with GFP via a single inter-chromosomal mitotic recombination event. Such a concurrent gene inactivation and cell labeling allows the study of the tumor initiation within hours of TSG loss. MADM also provides a perfect internal control, RFP labeled WT sibling cells of the GFP labeled mutant cells. Therefore, our lab will use the MADM system to model LOH of tumor suppressor genes (TSGs) and to study the detailed mechanisms of tumor initiation and progression.
How does MADM do the trick?
Click here to: View the presentation on MADM
- The basic components of MADM is a pair of chimeric fluorescent protein genes that are knocked-in at an equivalent locus of homologous chromosomes. The animal will be colorless because of the chimera proteins do not code functional proteins.
- MADM does not knock-out genes. A pre-existing mutated gene at the telomeric side needs to be recombined onto one of the MADM-bearing chromosomes. So our starting material is a heterozygous, colorless animal.
- MADM makes use of mitotic recombination to generate homozygous mutant cells. A single event of Cre-loxP mediated recombination will result in the reconstitution of fluorescent proteins and the movement of the mutant allele.
- Through Z segregation, MADM will generate a yellow, heterozygous cell that is not very informative. However, a X segregation will result in a green mutant cell, and a red sibling wildtype cell perfect internal control).
- All progeny from these colored cells will inherit the color, allowing MADM to be used as a lineage tracing tool. We hope to take advantage of this feature to locate cell-of-origin of cancers, and open the black box of tumor initiation and early progression mechanisms.
- Please see Zong et al, Cell 2005 for details.
Current Projects
Medulloblastoma modeling
 Medulloblastoma is the most common malignant brain tumor in children, and resides in the cerebellum of affected patients. Current therapy involves cytotoxic agents, including radiation and chemotherapy. Though partially effective, these methods are not completely satisfactory because of their long-term detrimental effects, in particular on the normal development of young patients. Therefore, it is very important to understand the molecular mechanisms of medulloblastoma, which will allow the development of more specific and less toxic drugs to improve the treatment efficacy. Clinical and human genetic studies have indicated that multiple genetic alterations are involved in the formation of medulloblastoma. In particular, both a deregulation of Shh signaling pathway and a defective DNA repair machinery could contribute to the tumorigenesis.
Medulloblastoma is the most common malignant brain tumor in children, and resides in the cerebellum of affected patients. Current therapy involves cytotoxic agents, including radiation and chemotherapy. Though partially effective, these methods are not completely satisfactory because of their long-term detrimental effects, in particular on the normal development of young patients. Therefore, it is very important to understand the molecular mechanisms of medulloblastoma, which will allow the development of more specific and less toxic drugs to improve the treatment efficacy. Clinical and human genetic studies have indicated that multiple genetic alterations are involved in the formation of medulloblastoma. In particular, both a deregulation of Shh signaling pathway and a defective DNA repair machinery could contribute to the tumorigenesis.
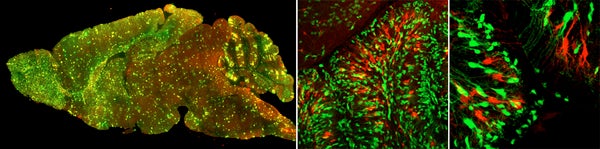
Recently, we established a medulloblastoma model with perturbations in both pathways, namely MADM-induced mosaic p53 loss and a Ptc heterozygous mutation background. Using this genetic combination, we have successfully generated a MADM-based medulloblastoma model. It is the first time anyone showed that tumors can be induced from mosaic TSG mutant mice, closely mimicking the clonal origin of human cancers. More importantly, tumors identified from these brains are entirely green, despite of very low number of green cells in unaffected areas. This observation strongly suggests that a tumor induced by MADM was originated from the clonal expansion of a single mutant cell. Using this model, we are studying the intricate interactions between tumor cells and tumor microenvironment (TME), with the ultimate goal to develop treatment strategies that destroy the supporting system for cancer to grow.
Glioma modeling
Glioma is the most common type of brain tumors in humans. The malignant form of glioma is incurable due to its elusive tumor origin, highly diffusive nature and complicated tumor organization. The hope for better treatment lies within the identification of the cell of origin of glioma, the depiction of tumor-niche interactions, and the delineation of molecular changes during tumorigenesis. Using MADM, we resolved a long-held controversy in the glioma field – which is the cell of origin, neural stem cells (NSCs) or oligodendrocyte precursor cells (OPCs)? While mutations introduced into NSCs could lead to gliomagenesis, traditional mouse models wouldn’t allow the analysis of the tumorigenic process, thus can’t rule out the possibility of downstream progenitor cells as the true cell of origin. MADM-based glioma model allowed us to examine mutant NSCs and all the progeny cell lineages at pre-malignant stages, and revealed significant aberrant growth only in the oligodendrocyte precursor cell (OPC) lineage. Our findings demonstrated that OPCs serve as the cell-of-origin for glioma even when initial mutations occur in NSCs, and highlight the importance of analyzing early phases of tumorigenesis to distinguish cell-of-origin from cell-of-mutation: while cell-of-mutation could be as early as in germline cells, only the cell type with susceptible signaling context can serve as the cell-of-origin to manifest malignant transformation [Liu 2011 Cell]. After the publication of our paper, the analytical principles have been adopted by many labs, and are expected to have a long-lasting impact in the field. After the initial work, we found that mutant OPCs massively outcompete WT OPCs long before malignancy, which is critical for tumor progression. Currently, we are studying the molecular mechanisms of OPC competition with the ultimate goal to stop glioma’s destructive force.
Future outlook
- Expand MADM-based cancer modeling to BRCA1-related breast and ovarian cancer.
- Apply MADM cancer models to pre-clinical drug tests.
- Live imaging to study tumor-TME interactions & metastasis.