Publications
SELECTED PUBLICATIONS
*Equal Contribution, # Corresponding Author
 1. “Ribosomes Hibernate on the Mitochondria during Cellular Stress. “
1. “Ribosomes Hibernate on the Mitochondria during Cellular Stress. “
Gemin,O.*, Gluc, M.*, Rosa, H, Purdy, M, Niemann, M, Peskova Y, Mattei, S,# Jomaa, A# 2024. Nat Comms. Oct 8;15(1):8666.
 2. “The translating bacterial ribosome at 1.55 Å resolution generated by cryo-EM imaging services”
2. “The translating bacterial ribosome at 1.55 Å resolution generated by cryo-EM imaging services”
Fromm* SA, O’Connor KM, Purdy M, Bhatt PR, Loughran G, Atkins# JF, Jomaa*# A, Mattei# S. 2023. Nat Comms. Feb 25;14(1):1095.
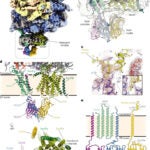 3. “Molecular basis of the TRAP complex function in ER protein biogenesis.”
3. “Molecular basis of the TRAP complex function in ER protein biogenesis.”
Jaskolowski*, M, Jomaa*#, A, Gamerdinger*, M, Shrestha, S, Leibundgut, M, Deuerling#, E, Ban#. N, 2023. Nat Struct Mol Biol. May 11. doi: 10.1038/s41594-023-00990-0
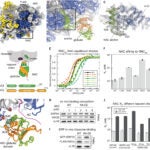 4. “Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.”
4. “Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.”
Jomaa*, A,Gamerdinger*, M, Hsieh*, H, Wallisch, A, Chandrasekaran, V, Ulsoy, Z, Scaiola, A, Hegde, RS, Shan#, S, Ban#, N, Deuerling#, E. 2022. Science. Feb 25;375(6583):839-844.
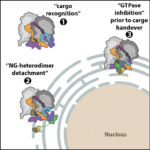 5. “Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.”
5. “Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.”
Jomaa, A#, Eitzinger, S, Zhu, Z, Chandrasekar, S, Kobayashi, K, Shan#, S, Ban#, N. 2021. Cell Reports. Jul 13;36(2):109350. doi: 10.1016/j.celrep.2021.109350.
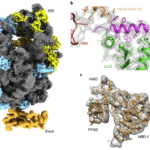 6. “Molecular mechanism of co-translational membrane protein recognition and targeting by SecA.”
6. “Molecular mechanism of co-translational membrane protein recognition and targeting by SecA.”
Wang*, S, Jomaa*, A, Jaskolowski, M, Yang, CI, Ban, N, Shan, S. 2019. Nat Struct Mol Biol. Oct;26(10):919-929.
7. “Structure of a pre-handover mammalian ribosomal SRP•SRP receptor targeting complex.”
Kobayashi*, K, Jomaa*, A, Lee, JH, Chandrasekar, S, Boehringer, D, Shan, So, Ban, N. 2018. Science. 2018 Apr 20;360(6386):323-327
 8. “Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.”
8. “Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.”
Jomaa, A, Hwang Fu, YH, Boehringer, D, Leibundgut, M, Shan, So, Ban, N. 2017. Nat Comm. May 19;8:15470.
CHRONOLOGICAL ORDER
*Equal Contribution
# Corresponding Author
(30) Zdancewicz, S., Maldosevic, E., Malezyna, K., Jomaa, A., NAC Couples Protein Synthesis with Nascent Polypeptide Myristoylation on the Ribosome. 2025, The EMBO Journal. https://doi.org/10.1038/s44318-025-00548-4
(29) Ren Q, Wang J, Idikuda V, Zhang S, Shin J, Ludlam WG, Real Hernandez LM, Zdancewicz S, Kreutzberger AJB, Chang H, Kiessling V, Tamm LK, Jomaa A, Levental I, Martemyanov K, Chanda B, Bao H. DeFrND: detergent-free reconstitution into native nanodiscs with designer membrane scaffold peptides. Nat Commun. 2025;16:7973. doi:10.1038/s41467-025-63275-8.
(28) Maldosevic, E*., Gora, R*, Lin, LL, Zhou, LE, Li, ZJ, Peskova, Y, Qi, L, Shan, S*, Jomaa, A*, A molecular switch in NAC prevents mitochondrial protein mistargeting by SRP, 2025 Biorxiv
(27) Lin, L#, Maldosevic, E, Zhou, L, Jomaa, A#, Qi, L#,Structural basis and pathological implications of the dimeric OS9-SEL1L-HRD1 ERAD Core Complex , 2025 Biorxiv
(26) Gemin,O.*, Gluc, M.*, Rosa, H, Purdy, M, Niemann, M, Peskova Y, Mattei, S,# Jomaa, A# Ribosomes Hibernate on the Mitochondria during Cellular Stress. Nat Commun. 2024 Oct 8;15(1):8666. doi: 10.1038/s41467-024-52911-4. PubMed PMID: 39379376.
(25) Cell cycle specific, differentially tagged ribosomal proteins to measure phase specific transcriptomes from asynchronously cycling cells. Sci Rep. 2024 Jan 18;14(1):1623. doi: 10.1038/s41598-024-52085-5.
(24) NAC controls cotranslational N-terminal methionine excision in eukaryotes. Science. 2023 Jun 23;380(6651):1238-1243. doi: 10.1126/science.adg3297. Epub 2023 Jun 22. PubMed PMID: 37347872.
(23) Jaskolowski*, M, Jomaa*#, A, Gamerdinger*, M, Shrestha, S, Leibundgut, M, Deuerling#, E, Ban#. N,2023. Molecular basis of the TRAP complex function in ER protein biogenesis.Nat Struct Mol Biol. May 11. doi: 10.1038/s41594-023-00990-0
(22) Fromm* SA, O’Connor KM, Purdy M, Bhatt PR, Loughran G, Atkins# JF, Jomaa*# A, Mattei# S. 2023. The translating bacterial ribosome at 1.55 Å resolution generated by cryo-EM imaging services. Nat Comms. Feb 25;14(1):1095.
(21) Jomaa*, A,Gamerdinger*, M, Hsieh*, H, Wallisch, A, Chandrasekaran, V, Ulsoy, Z, Scaiola, A, Hegde, RS, Shan#, S, Ban#, N, Deuerling#, E. 2022. Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting. Science.Feb 25;375(6583):839-844.
(20) Kavalchuk*, M, Jomaa*#, A, Müller, AU, Weber-Ban#, E. 2022 Structural basis of prokaryotic ubiquitin-like protein engagement and translocation by the mycobacterial Mpa-proteasome complex. Nat. Comms. Jan 12;13(1):276. doi: 10.1038/s41467-021-27787-3.
(19) Jomaa#, A, Eitzinger, S, Zhu, Z, Chandrasekar, S, Kobayashi, K, Shan#, S, Ban#, N. 2021. Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle. Cell Reports. Jul 13;36(2):109350. doi: 10.1016/j.celrep.2021.109350.
(18) Lee*, JH, Jomaa*#, A, Chung, S, Hwang Fu YH, Qian, R, Sun, X, Chandrasekar S, Hsieh, HH, Bi, X, Mattei, S, Boehringer, D, Weiss, S, Ban#, N, Shan#, S. 2021. Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER. Science Advances. May 21;7(21):eabg0942. doi: 10.1126/sciadv.abg0942.
(17) Schubert*, K, Karousis*, ED, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, LA, Leibundgut, M, Thiel, V, Mühlemann, O, Ban, N. 2020. SARS-CoV-2 Nsp1 binds ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. Sep 9. doi: 10.1038/s41594-020-0511-8.
(16) Wang*, S, Jomaa*, A, Jaskolowski, M, Yang, CI, Ban, N, Shan, S. 2019. Molecular mechanism of co-translational membrane protein recognition and targeting by SecA. Nat Struct Mol Biol. Oct;26(10):919-929. (Recommended in Faculty of 1000 Prime)
(15) Gamerdinger*, M, Kobayashi*, K, Wallisch, A, Kreft, SG, Sailer, C, Schlömer, R, Sachs, N, Jomaa, A, Stengel, Ban, N, Deuerling, E. 2019. Early scanning of nascent polypeptides inside the ribosomal tunnel by NAC. Mol Cell. Sep 5;75(5):996-1006.
(14) Ziemski M, Jomaa, A, Mayer, D, Rutz, S, Giese, C, Verpintsev, D, Weber-Ban, E. 2018 Cdc48-like protein of actinobacteria (Cpa) is a novel proteasome interactor in mycobacteria and related actinobacteria. Elife. 2018 May 29;7. pii: e34055
(13) Kobayashi*, K, Jomaa*, A, Lee, JH, Chandrasekar, S, Boehringer, D, Shan, So, Ban, N. 2018. Structure of a pre-handover mammalian ribosomal SRP•SRP receptor targeting complex. Science. 2018 Apr 20;360(6386):323-327 (Recommended in Faculty of 1000 Prime)
(12) Jomaa, A, Hwang Fu, YH, Boehringer, D, Leibundgut, M, Shan, So, Ban, N. 2017. Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome. Nat Comm. May 19;8:15470.
(11) Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. 2016. Structures of the E. coli translating ribosome with the SRP and its receptor and with the translocon. Nat Comm. Jan 25;7:10471. (Recommended in Faculty of 1000 Prime)
-
During graduate studies:
(10) Jomaa *, A, Jain* N, Davis* JH, Williamson, JR, Britton, RA, Ortega J. 2014. Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Res. Mar;42(5):3419-35.
(9) Chen, C, Jomaa, A, Ortega, J, and Alani, EE. 2014. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc Natl Acad Sci U S A. Jan 7;111(1):E44-53.
(8) Leong, V, Kent, M, Jomaa, A, Ortega J. 2013. Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement. RNA. Jun;19(6):789-802.
(7) Jomaa, A, Stewart G, Mears, JA, Kireeva, I, Brown, ED, Ortega J. 2011. Cryoelectron microscopy structure of the 30S subunit in complex with the YjeQ biogenesis factor. RNA. Nov;17(11):2026-38. (Featured on the journal’s cover page)
(6) Capstick,DS, Jomaa, A, Hanke, C, Ortega, J, and Elliot MA 2011. Dual amyloid domains promote differential functioning of the chaplin proteins during Streptomyces aerial morphogenesis. Proc Natl Acad Sci U S A. Jun 14;108(24):9821-6.
(5) Jomaa, A, Stewart, G, Martín-Benito, J, Zielke, R., Campbell, TL, Maddock JR, Brown, E.D., J. Ortega. 2011. Understanding Ribosome Assembly: the Structure of In vivo Assembled Immature 30S Subunits Revealed by Cryo-electron Microscopy. RNA. Apr;17(4):697-709.
-
During undergraduate studies:
(4) Ortega, J, Iwanczyk, J, Jomaa, A. 2009. Escherichia coli DegP: a structure driven functional model. J Bacteriol. Aug;191:4705-13. (Mini-review)
(3) Jomaa, A, Iwanczyk, J, Tran, J, Ortega, J, 2009. Characterization of the autocleavage process of the Escherichia coli HtrA protein: implications for its physiological role. J Bacteriol. Mar; 191:1924-32. (Featured on the journal’s cover page)
(2) Iwanczyk, J, Damjanovic, D, Kooistra, J., Leong, V., Jomaa, A, Ghirlando, R., Ortega, J. 2007. Role of the PDZ domains in Escherichia coli DegP Protein. J. Bacteriol. Apr; 189:3176-3186.
(1) Jomaa, A, Damjanovic, D, Leong, V, Ghirlando, R, Iwanczyk, J, Ortega, J. 2007. The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity. J. Bacteriol. Feb; 189:706-716. (Featured on the journal’s cover page).
