Zhang Lab
Genome Integrity Maintenance within Repetitive Sequences
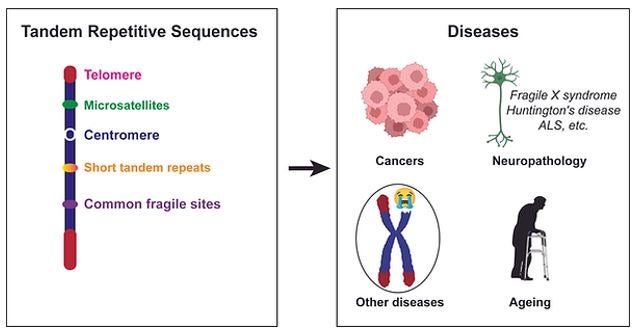
Tandem repetitive sequences, such as telomere, centromere, rDNA, short tandem repeats like (CGG)n or (TA)n, etc., can form a unique chromatin environment with complex secondary structures. The genome instability within tandem repetitive sequences leads to multiple diseases, including cancers, neuropathological diseases, and ageing-related syndromes. The highly repetitive nature of these genome regions makes it difficult to analyze. Tandem repetitive sequences thus have long been recognized as the dark side of the human genome. Tandem repetitive sequences are characterized as difficult-to-replicate regions and DNA damage hotspots. The genetic and epigenetic regulation of DNA damage responses within tandem repetitive sequences is a fascinating yet largely enigmatic subject. We are dedicated to unraveling how DNA replication and repair through the complex secondary structures in the tandem repetitive sequences and shedding light on their associated etiology and therapeutic vulnerabilities.
 Tianpeng Zhang, Ph.D.
Tianpeng Zhang, Ph.D.
Assistant Professor
Department of Radiation Oncology
Department of Biochemistry & Molecular Genetics
University of Virginia School of Medicine
Lab Room 7111, Multistory Building, West Complex, PO Box 800383
Lab: 434-243-1398
Office: 434-243-1398
Lab Website: www.ztplab.com
Email: zjv8au@virginia.edu
Publications
- Jiang HY*, Zhang TP*, Kaur H, Shi T, Krishnan A, Kwon Y, Sung P, and Greenberg RA. (2024). BLM helicase unwinds lagging strand flap substrates to assemble the ALT telomere damage response. Mol Cell 84, 1684-1698 e1689. (* equal contribution; Cover story).
- Zhang TP, Rawal Y, Jiang HY, Kwon Y, Sung P and Greenberg RA. (2023). Break-induced replication orchestrates resection-dependent template switching. Nature 619, 201-208.
- Zhang TP, Greenberg RA. (2022). The inner workings of replisome-dependent control of DNA damage tolerance. Genes Dev 36 (3-4), 103-105 (Preview)
- Vessoni AT, Zhang TP, Quinet A, Jeong HC, Munroe M, Wood M, Tedone E, Vindigni A, Shay JW, Greenberg RA, Batista LFZ. (2021). Telomere erosion in human pluripotent stem cells leads to ATR-mediated mitotic catastrophe. J Cell Biol 220(6): e202011014.
- Hu JL, Liang H, Zhang H, Yang MZ, Sun W, Zhang P, Luo L, Feng JX, Bai HJ, Liu F, Zhang TP, Yang JY, Gao QS, Long YK, Ma XY, Chen Y, Zhong Q, Yu B, Liao S, Wang YB, Zhao Y, Zeng MS, Cao N, Wang JC, Chen W, Yang HT, Gao S. (2020). FAM46B is a prokaryotic-like cytoplasmic poly (A) polymerase essential in human embryonic stem cells. Nucleic Acids Res 48, 2733-2748.
- Zhang ZP*, Zhang TP*, Ge YL*, Tang MF, Ma WB, Zhang QF, Gong SZ, Wright WE, Shay JW, Liu HY and Zhao Y. (2019). 2D gel electrophoresis reveals dynamics of t-loop formation during the cell cycle and t-loop in maintenance regulated by heterochromatin state. J Biol Chem 294, 6645-6656. (* equal contribution)
- Zhang TP*, Zhang ZP*, Gong SZ, Li XC, Liu HY and Zhao Y. (2019). Strand break-induced replication fork collapse leads to C-circles, C-overhangs and telomeric recombination. Plos Genet 15 (2), e1007825. (* equal contribution)
- Verma P, Dilley RL*, Zhang TP*, Gyparaki MT, Li YW and Greenberg RA. (2019). Rad52 and SLX4 Act non-epistatically to ensure telomere stability during Alternative Telomere Lengthening. Genes Dev, 33, 221- (* equal contribution)
- Li F, Kim H, Ji ZJ, Zhang TP, Chen BH, Ge YL, Hu Y, Feng XY, Han X, Xu HM, Zhang YW, Yu HT, Liu D, Ma WB and Songyang Z. (2018). The BUB3-BUB1 complex promotes telomere DNA replication. Mol Cell 70, 395-407.
- Zhang TP, Zhang ZP, Li F, Hu Q, Liu HY, Tang MF, Ma WB, Huang JJ, Songyang Z, Rong YK, Zhang SC, Chen BP and Zhao Y. (2017). Looping-out mechanism for resolution of replication stress at telomeres. EMBO Rep 18, 1412-1428.
- Liu HY, Zhao Q, Zhang TP, Wu Y, Xiong YX, Wang SK, Ge YL, He JH, Lv P, Ou TM, Tan JH, Li D, Gu LQ, Ren J, Zhao Y and Huang ZS. (2016). Conformation selective antibody enables genome profiling and leads to discovery of parallel G-quadruplex in human telomeres. Cell Chem Biol 23, 1261-1270.
- Ren XY*, Yang XJ*, Cheng B*, Chen XZ*, Zhang TP, He QM, Li B, Li YQ, Tang XR, Wen X, Zhong Q, Kang TB, Zeng MS, Liu N and Ma J. (2017). HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal. Nature Commun 8, 14053.
- Xie C*, Chen YL*, Wang DF, Wang YL, Zhang TP, Li H, Liang F, Zhao Y, and Zhang GY. (2017). SgRNA Expression of CRIPSR-Cas9 system based on miRNA polycistrons as a versatile tool to manipulate multiple and tissue-specific genome editing. Sci Rep 7, 5795. (* equal contribution)
- Zheng XH, Mu G, Zhong YF, Zhang TP, Cao Q, Ji LN, Zhao Y and Mao ZW. (2016). Trigeminal star-like platinum complexes induce cancer cell senescence through quadruplex-mediated telomere dysfunction. Chem Commun (Camb) 52, 14101-14104.
- Wu SH, Zhang TP and Du JS. (2016). Ursolic acid sensitizes cisplatin-resistant HepG2/DDP cells to cisplatin via inhibiting Nrf2/ARE pathway. Drug Des Devel Ther 10, 3471-3481.
