Membrane Proteins
Structure-Function-Dynamics-Folding of Membrane Proteins
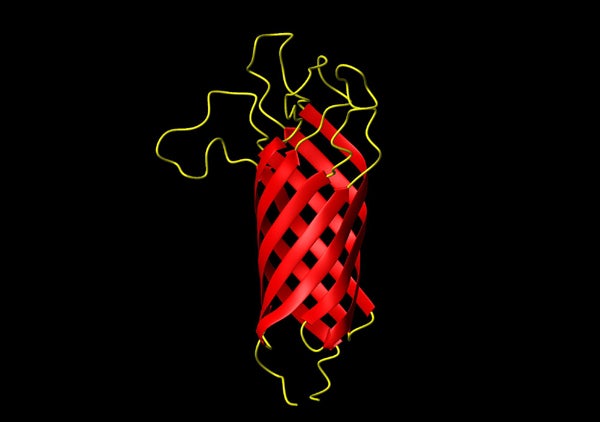
Membrane proteins – receptors, ion channels, transporters, etc. – constitute about 30% of all proteins in eukaryotic cells. Many of them are targets for current or future drugs. In order to facilitate the basic understanding of the biology of these proteins and in order to aid future rational drug design improved methods are needed to solve the structures of this class of proteins. Since membrane proteins are harder to express and handle than soluble proteins, specialized expression, solubilization, and stabilization techniques are required. Our laboratory is active in all these areas and we are particularly interested in solving structures of membrane proteins by solution NMR spectroscopy. We have determined the very first membrane protein structure that was solved by this technique in 2001, i.e. that of the outer membrane ion channel OmpA.
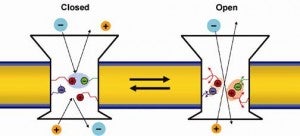
We also performed dynamic, thermodynamic, and electrophysiological single channel recording experiments to delineate the gating mechanism of this ion channel.
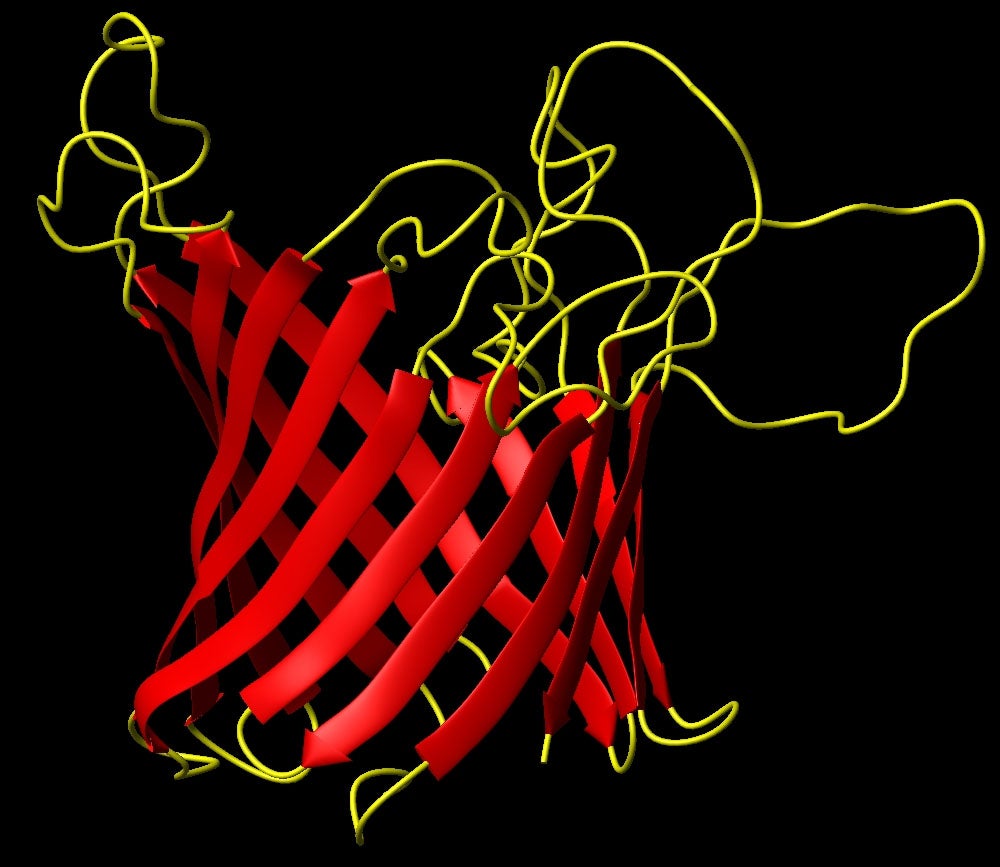
In 2007 we determined the structure of the outer membrane porin OmpG by NMR spectroscopy. This is currently one of the largest membrane protein structures ever solved by NMR, (33 kDa, 280 residues).
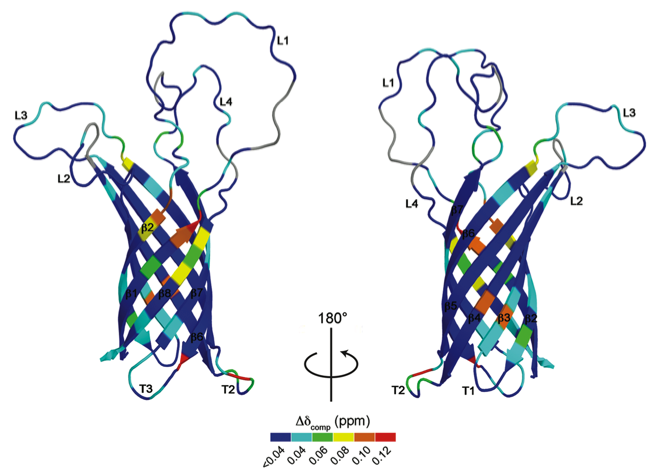
More recently, we solved the structures of OprH and OprG from Pseudomonas aeruginosa. We found that OprG is a facilitator for small amino acid transport in the outer membrane of this clinically important pathogen. We also determined the interaction of OprH with lipopolysaccharide (LPS) by NMR. The tight OprH-LPS interaction has been implicated in forming an unusually tough outer membrane and thereby conferring antibiotic resistance. These studies will help us to understand and improve antibiotic treatment of Pseudomonas infections.
The methods that we are developing with these proteins should also be helpful to tackle more difficult projects in the future like helical membrane receptors and ion channels that have been identified as potential drug targets.
- Structure of E.coli OmpA
- Gating of OmpA Ion Channel
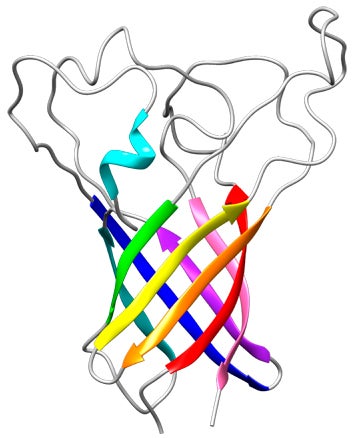
- Structure of E. coli OmpG
- Structure of P. aeruginosa OprH
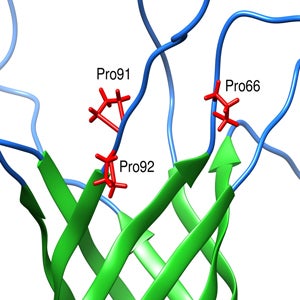
- Proline rich region in NMR structure of P. aeruginosa OprG contributes to transport of small amino acids across outer membrane
- Structure of P. aeruginosa OprG
**click to view full size image
Recent Key Publications:
Crawford MA, Ward AE, Gray V, Bailer P, Fisher DJ, Kubicka E, Cui Z, Luo Q, Gray MC, Criss AK, Lum LG, Tamm LK, Letteri RA, Hughes MA. (2023) Disparate regions of the human chemokine CXCL10 exhibit broad-spectrum antimicrobial activity against biodefense and antibiotic-resistant bacterial pathogens. ACS Infect Dis 9:122-139. (https://pubmed.ncbi.nlm.nih.gov/36475632/)
Vorobieva AA, White P, Liang B, Horne JE, Bera AK, Chow CM, Gerben S, Marx S, Kang A, Stiving AQ, Harvey SR, Marx DC, Khan GN, Fleming KG, Wysocki VH, Brockwell DJ, Tamm LK, Radford SE, Baker D (2021) De novo design of transmembrane β-barrels. Science 371, 801. doi: 10.1126/science.abc8182. (https://pubmed.ncbi.nlm.nih.gov/33602829/)
Sanganna Gari RR, Seelheim P, Liang B, Tamm LK. Quiet Outer Membrane Protein G (OmpG) Nanopore for Biosensing. ACS Sens. 2019 May 24;4(5):1230-1235. doi: 10.1021/acssensors.8b01645. Epub 2019 Apr 25. (https://www.ncbi.nlm.nih.gov/pubmed/30990011)
Sanganna Gari RR, Seelheim P, Marsh B, Kiessling V, Creutz CE, Tamm LK. Quaternary structure of the small amino acid transporter OprG from Pseudomonas aeruginosa. J Biol Chem. 2018 Nov 2;293(44):17267-17277. doi: 10.1074/jbc.RA118.004461. Epub 2018 Sep 20. (https://www.ncbi.nlm.nih.gov/pubmed/30237175)
Blackburn MR, Hubbard C, Kiessling V, Bi Y, Kloss B, Tamm LK, Zimmer J. Distinct reaction mechanisms for hyaluronan biosynthesis in different kingdoms of life. Glycobiology. 2018 Feb 1;28(2):108-121. doi: 10.1093/glycob/cwx096. (https://www.ncbi.nlm.nih.gov/pubmed/29190396)
Kucharska I, Tamm LK. Solution NMR Provides New Insight into Lipid-Protein Interaction. Biochemistry. 2017 Aug 22; 56(33): 4291–4292. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5568481/)
Chiu YH, Jin X, Medina CB, Leonhardt SA, Kiessling V, Bennett BC, Shu S, Tamm LK, Yeager M, Ravichandran KS, Bayliss DA. (2017) A quantized mechanism for activation of pannexin channels. Nat Commun. 2017 Jan 30;8:14324. (https://www.ncbi.nlm.nih.gov/pubmed/28134257)
Lee J, Patel DS, Kucharska I, Tamm LK, Im W. (2017) Refinement of OprH-LPS Interactions by Molecular Simulations. Biophys J. 2017 Jan 24;112(2):346-355. (https://www.ncbi.nlm.nih.gov/pubmed/28122220)
Kucharska I, Liang B, Ursini N, Tamm LK. (2016) Molecular Interactions of Lipopolysaccharide with an Outer Membrane Protein from Pseudomonas aeruginosa Probed by Solution NMR. Biochemistry. 2016 Sep 13;55(36):5061-72. (https://www.ncbi.nlm.nih.gov/pubmed/27532487)
Liang B, Tamm LK. (2016) NMR as a tool to investigate the structure, dynamics and function of membrane proteins. Nat Struct Mol Biol. 2016 Jun 7;23(6):468-74. (https://www.ncbi.nlm.nih.gov/pubmed/27273629)
Kucharska, I., Seelheim, P., Edrington, T., Liang, B., Tamm, L.K. (2015) OprG Harnesses the Dynamics of its Extracellular Loops to Transport Small Amino Acids across the Outer Membrane of Pseudomonas aeruginosa. Structure Dec. 1;23:2234-2245 (http://www.ncbi.nlm.nih.gov/pubmed/26655471)
Kucharska, I., Edrington, T.C., Liang, B., Tamm, L.K. (2015) Optimizing nanodiscs and bicelles for solution NMR studies of two Beta-barrel membrane proteins. J. Biomol NMR 61(3-4):261-274. (http://www.ncbi.nlm.nih.gov/pubmed/25869397)
Zhuang, T., Tamm, L. K. (2014) Control of the conductance of engineered protein nanopores through concerted loop motions. Angewandte Chemie. Jun 2;53(23):5897-902. (http://www.ncbi.nlm.nih.gov/pubmed/24777684)
Marcoux, J., Politis, A., Rinehart, D., Marshall, D.P., Wallace, M.I., Tamm, L.K., Robinson, C.V. (2014) Mass spectrometry defines the C-terminal dimerization domain and enables modeling of the structure of full-length OmpA. Structure May 6;22(5):781-90. (http://www.ncbi.nlm.nih.gov/pubmed/24746938)
Zhuang, T., Chisholm, C., Chen, M., and Tamm, L.K. (2013) NMR-based conformational ensembles explain pH-dependent opening and closing of OmpG channel. J. Am. Chem. Soc. Oct 9;135(40):15101-13. (http://www.ncbi.nlm.nih.gov/pubmed/24020969)
Hong H, Rinehart D, Tamm LK. (2013) Membrane depth-dependent energetic contribution of the tryptophan side chain to the stability of integral membrane proteins. Biochemistry. Jun 25;52(25):4413-21. (http://www.ncbi.nlm.nih.gov/pubmed/23763479)
Edrington, T.C. Kintz, E., Goldberg, J.B. and Tamm, L.K. (2011). Structural Basis for the Interaction of Lipopolysaccharide with the Outer Membrane Protein OprH from Pseudomonas Aeruginosa. Jour. of Biol Chem. 286, 39211-39223. (http://www.ncbi.nlm.nih.gov/pubmed/21865172)
Liang, B., Arora, A., Tamm, L.K. (2010) Fast-time scale dynamics of Outer membrane protein A by extended model-free analysis of NMR relaxation data Biochim. Biophys. Acta, 1798, 68-76. (http://www.ncbi.nlm.nih.gov/pubmed/19665446)
Hong, H., Joh, N.H., Bowie, J.U., and Tamm, L.K. (2009) Methods for Measuring the Thermodynamic Stability of Membrane Proteins. Methods in Enzymology. Vol. 455: 213-236. (http://www.ncbi.nlm.nih.gov/pubmed/19289208)
Liang B. and Tamm, L.K. (2007) Structure of outer membrane protein G by solution NMR spectrscopy. PNAS. Vol. 104 no.41:16140-16145. (http://www.ncbi.nlm.nih.gov/pubmed/17911261)
Hong, H., Park, S., Flores-Jiménez, R.H., Rinehart, D., and Tamm, L.K. (2007) Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J.A.C.S. 129:8320-8327. (http://www.ncbi.nlm.nih.gov/pubmed/17564441)
Hong H., Szabo G., Tamm L.K. (2006) Electrostatic Coupling in OmpA Ion-Channal Gating Suggest a Mechanism for Pore Opening. Nature Chem. Biol. Vol.2 No.11 627-635. (http://www.ncbi.nlm.nih.gov/pubmed/17041590)
Tamm L.K., Liang B. (2006). NMR of membrane protein in solution. Prog. in Nuc Mag Res Spec. Vol.48 201-210.
Cierpicki T., Liang B., Tamm L.K., Bushweller J.H.. (2006). Increasing the Accuracy of Solution NMR Structures of Membrane Proteins by Application of Residual Dipolar Coupling. High-Resolution Structure of Outer Membrane Protein A.. J. Am Chem. Soc Vol.128 No.21 6947-6951. (http://www.ncbi.nlm.nih.gov/pubmed/16719475).
Liang B., Bushweller J.H., Tamm L.K. (2006). Site-Directed Parallel Spin-Labeling and Paramagnetic Relaxation Enhancement in Structure Determination of Membrane Proteins by Solution NMR Spectroscopy. J. Am Chem. Soc Vol.128 No.13 4389-4397. (http://www.ncbi.nlm.nih.gov/pubmed/16569016).
Hong, H., Patel D.R., Tamm L.K. and Berg B.V.D. (2006). The Outer Membrane Protein OmpW Forms an Eight-stranded β-Barrel with a Hydrophobic Channel. Jour. of Biol Chem. Vol. 281 No.11: 7568-7577. (http://www.ncbi.nlm.nih.gov/pubmed/16414958).
Tamm, L.K., H. Hong, and B. Liang (2004). Folding and assembly of beta-barrel membrane proteins. (Review) Biochim. Biophys. Acta. 1666: 250-263. (http://www.ncbi.nlm.nih.gov/pubmed/15519319).
Hong, H. and L.K. Tamm (2004). Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Nat. Acad. Sci. 101: 4065-4070 (http://www.ncbi.nlm.nih.gov/pubmed/14990786). Commentary by Dr. James Bowie.
Tamm, L.K., F. Abildgaard, A. Arora, H. Blad, and J.H. Bushweller (2003). Structure, dynamics and function of the outer membrane protein A (OmpA) and influenza hemagglutinin fusion domain in detergent micelles by solution NMR. (Minireview) FEBS Lett. 555: 139-143. (http://www.ncbi.nlm.nih.gov/pubmed/14630334)
Kleinschmidt, J.H. and L.K. Tamm (2002). Secondary and tertiary structure formation of the beta-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J. Mol. Biol. 324: 319-330. (http://www.ncbi.nlm.nih.gov/pubmed/12441110)
Kleinschmidt, J.H. and L.K. Tamm (2002). Structural transitions in short-chain lipid assemblies studied by P(31)-NMR spectroscopy. Biophys. J. 83: 994-1003. (http://www.ncbi.nlm.nih.gov/pubmed/12124281)
Arora, A. and L.K. Tamm (2001). Biophysical approaches to membrane protein structure determination. (Review) Curr. Opin. Struct. Biol. 11: 540-547. (http://www.ncbi.nlm.nih.gov/pubmed/11785753)
Tamm, L.K., A. Arora, and J.H. Kleinschmidt (2001). Structure and assembly of beta-barrel membrane proteins. (Review) J. Biol. Chem. 276: 32399-32402. (http://www.ncbi.nlm.nih.gov/pubmed/11432877)
Arora, A., A. Frits, J.H. Bushweller, and L.K. Tamm (2001). Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nature Struct. Biol. 8: 334-338. (http://www.ncbi.nlm.nih.gov/pubmed/11276254)