Virus Fusion
Membrane Fusion in Viral Infection
Our laboratory is interested in unraveling the mechanisms of membrane fusion when enveloped viruses such as influenza, HIV, or Ebola enter their respective host cells. To do so, we take combined structural, biophysical, and cell biological approaches. Ultimately we want to understand how triggered fusion proteins deform their own and the host cells’ membranes so that they merge into a single membrane and thereby deliver the viral genome into the cell. Understanding the fundamental biological underpinnings of this process will facilitate the discovery of new antiviral therapies.
- Flu virus particle
- Human Immunodeficiency Virus Particle
- Electron micrograph of Ebola Virus
Although the structures of the soluble domains of many viral fusion proteins have been solved, the mechanism of membrane fusion is still poorly understood. We are studying the structures and interactions of viral fusion proteins in lipid bilayers. We have worked out a detailed “spring-loaded boomerang” model of how influenza HA interacts with viral and cellular membranes.
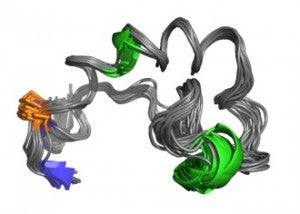
We have determined by NMR and spin-label EPR spectroscopy, the structures of the influenza HA fusion domain in lipid bilayers at resting and fusion pH. We also studied many mutants which allowed us to establish structural features that are critical for proper function of these important viral protein domains.
More recently we have engaged in similar studies with the fusion domain of HIV gp41 and, in collaboration with Judith White’s laboratory, with the internal loop fusion domain of Ebola virus GP2.
We have also studied how HIV particles bind to and fuse at the boundaries between raft and non-raft regions of model and cellular membranes.
We recently received a competitive multi-investigator research grant from the Human Frontier Science Program (HFSP) to study changes in the properties of influenza membrane envelopes when avian flu strains are transmitted from avian to human cell growth conditions. The other investigators in this consortium are Michael Veit (Free University of Berlin), Markus Wenk (National University of Singapore), and Kay Grünewald (University of Oxford).
- HIV virosomes bind and fuse to lipid model membranes at the edges of lipid rafts
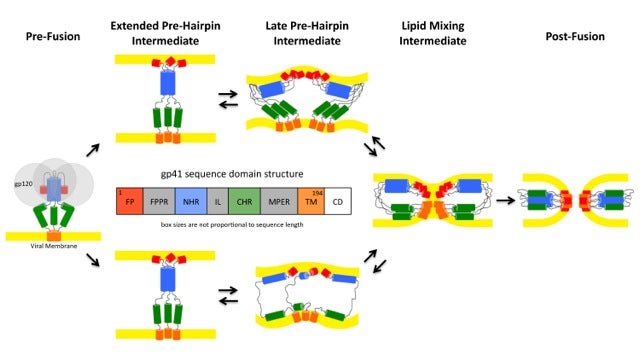
- Pathways of conformational intermediates in HIV gp41-mediated membrane fusion
- Structures of influenza virus fusion peptides in lipid membranes.
- Structure of Ebola virus fusion loop.
Recent Key Publications
White JM, Ward AE, Odongo L, Tamm LK (2023). Viral membrane fusion: A dance between proteins and lipids. Ann Rev Virology 10:139-161.
Odongo L, Habtegebrael B, Kiessling VK, White JM, Tamm LK (2023) A novel in vitro system of supported planar endosomal membranes (SPEMs) reveals an enhancing role for cathepsin B in the final stage of Ebola virus fusion and entry. Microbiology Spectrum e0190823. doi: 10.1128/spectrum.01908-23.
Ward AE, Sokovikova D, Waxham MN, Heberle FA, Levental I, Levental KR, Kiessling V, White JM, Tamm LK (2023) Serinc5 restricts HIV membrane fusion by altering lipid order and heterogeneity in the viral membrane. ACS Infect Dis DOI: 10.1021/acsinfecdis.2c00478
Odongo L, Zadrozny KK, Diehl WE, Luban L, White JM, Ganser-Pornillos BK, Tamm LK, Pornillos O (2023). Purification and structure of luminal domain C of human Niemann-Pick C1 protein. Acta Cryst. F79:45-50. (https://pubmed.ncbi.nlm.nih.gov/36748341/)
Cabot M, Kiessling V, White JM, Tamm LK (2022) Endosomes supporting fusion mediated by vesicular stomatitis virus glycoprotein have distinctive motion and acidification. Traffic 23:221-234. (https://pubmed.ncbi.nlm.nih.gov/35147273/)
Lee J, Kreutzberger AJB, Odongo L, Nelson EA, Nyenhuis DA, Kiessling V, Liang B, Cafiso DS, White JM, Tamm LK (2021) Ebola virus glycoprotein interacts with cholesterol to enhance membrane fusion and cell entry Affiliations. Nat Struct Mol Biol. 2021 Jan 18. doi: 10.1038/s41594-020-00548-4. Online ahead of print. (https://pubmed.ncbi.nlm.nih.gov/33462517/)
Ward A, Kiessling V, Pornillos OW, White JM, Ganser-Pornillos BK, Tamm LK (2020) HIV-Cell membrane bleb fusion intermediates are restricted by Serincs as revealed by cryo electron and TIRF microscopy. J Biol Chem 295:15183-15195. (Editor’s pick. On the cover) (https://pubmed.ncbi.nlm.nih.gov/32788212/)
Lee J, Nyenhuis D, Nelson EA, Cafiso DS, White JM, Tamm LK (2017) Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc Natl Acad Sci USA, Epub doi:10.1073/pnas.1708052114. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5617291/)
Yang ST, Kreutzberger AJB, Kiessling V, Ganser-Pornillos BK, White JM, Tamm LK. (2017) HIV virions sense plasma membrane heterogeneity for cell entry. Sci. Adv. Jun 2017: Vol. 3, no. 6, e1700338 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5489272/)
Yang ST, Kreutzberger AJ, Lee J, Kiessling V, Tamm LK. (2016) The role of cholesterol in membrane fusion. Chem Phys Lipids. 2016 Sep;199:136-43. doi: 10.1016/j.chemphyslip.2016.05.003. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4972649/)
Yang ST, Kiessling V, Tamm LK. (2016) Line tension at lipid phase boundaries as driving force for HIV fusion peptide-mediated fusion. Nat Commun. 2016 Apr 26;7:11401. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4853434/)
Lee, J., Gregory, S.M., Nelson, E.A., White, J.M., Tamm, L.K. (2016) The Roles of Histidines and Charged Residues as Potential Triggers of a Conformational Change in the Fusion Loop of Ebola Virus Glycoprotein. PLoS One. Mar 29;11(3):e0152527. doi: 10.1371/journal.pone.0152527. eCollection 2016. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4811418/)
Yang ST, Kiessling V, Simmons JA, White JM, Tamm LK. (2015) HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat Chem Biol. Jun;11(6):424-31. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4433777/)
Tamm LK, Lee J, Liang B. (2014) Capturing glimpses of an elusive HIV gp41 prehairpin fusion intermediate. Structure. Sep 2;22(9):1225-6 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4199205/)
Gregory, S.M., Larsson, P., Nelson, E.A., Kasson, P.M., White, J.M., Tamm, L.K. (2014) Ebolavirus Entry Requires a Compact Hydrophobic Fist at the Tip of the Fusion Loop. J. Virol Jun;88(12):6636-49. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4054381/)
Smith EC, Gregory SM, Tamm LK, Creamer TP, Dutch RE. (2012) Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion. J Biol Chem. Aug 24;287(35):30035-48. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3436143/)
Lai, A.L., Moorthy, A.E., Li, Y., Tamm, L.K. (2012) Fusion Activity of HIV gp41 Fusion Domain Is Related to Its Secondary Structure and Depth of Membrane Insertion in a Cholesterol-Dependent Fashion. Mol. Biol., Apr 20;418 ( https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3654243/)
Gregory, S.M., Harada, E., Liang, B., Delos, S.E., White, J.M., and Tamm, L.K. (2011). Structure and function of the complete internal fusion loop from ebolavirus glycoprotein 2. Proc. Natl. Acad. Sci. U.S.A. Jul 5;108(27):11211-6. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3131375/)
Lai, A.L. and Tamm, L.K. (2010) Shallow boomerang-shaped influenza hemagglutinin G13A mutant structure promotes leaky membrane fusion. J. Biol. Chem. Nov 26;285(48):37467-75. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2988352/)
Tamm, L.K., Lai, A.L., Li Y. (2007). Combined NMR and EPR spectroscopy to determine structures of viral fusion domains in membranes. Bioch. Biop. Acta. Dec;1768(12):3052-60. (http://www.ncbi.nlm.nih.gov/pubmed/17963720)
Lai, A.L. and Tamm, L.K. (2007) Locking the kink in the influenza hemagglutinin fusion domain structure. J. Biol. Chem. Aug 17;282(33):23946-56. (http://www.ncbi.nlm.nih.gov/pubmed/17567572)
Li, Y. and Tamm, L.K. (2007) Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophys. J. Aug 1;93(3):876-85. (http://www.ncbi.nlm.nih.gov/pubmed/17513369)
Lai AL, Park H, White JM, Tamm LK. (2006). Fusion Peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J Biol Chem. 2006 Mar 3;281(9):5760-70 (http://www.ncbi.nlm.nih.gov/pubmed/16407195)
Li, Y., Han,X., Lai, A. LQ, Bushweller J.H., Cafiso D.S., Tamm L.K. (2005). Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion. Journal of Virology Sep;79(18):12065-76 (http://www.ncbi.nlm.nih.gov/pubmed/16140782)
Tamm, L.K., J. Crane, and V. Kiessling (2003). Membrane fusion: a structural perspective on the interplay of lipids and proteins. (Review) Curr. Op. Struct. Biol. Aug;13(4):453-66 (http://www.ncbi.nlm.nih.gov/pubmed/12948775)
Tamm, L.K. (2003). Hypothesis: spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. (Review) Biochim. Biophys. Acta. Jul 11;1614(1):14-23. (http://www.ncbi.nlm.nih.gov/pubmed/12873762)
Li, Y., X. Han, and L.K. Tamm (2003). Thermodynamics of fusion peptide-membrane interactions. Biochemistry. Jun 17;42(23):7245-51. (http://www.ncbi.nlm.nih.gov/pubmed/12795621)
Han, X., J.H. Bushweller, D.S. Cafiso, and L.K. Tamm (2001). Membrane structure and fusion triggering conformational change of the fusion domain from influenza hemagglutinin. Nature Struct. Biol. Aug;8(8):715-20. (http://www.ncbi.nlm.nih.gov/pubmed/11473264)