Research
Mechanisms for Common Fragile Site Breakage
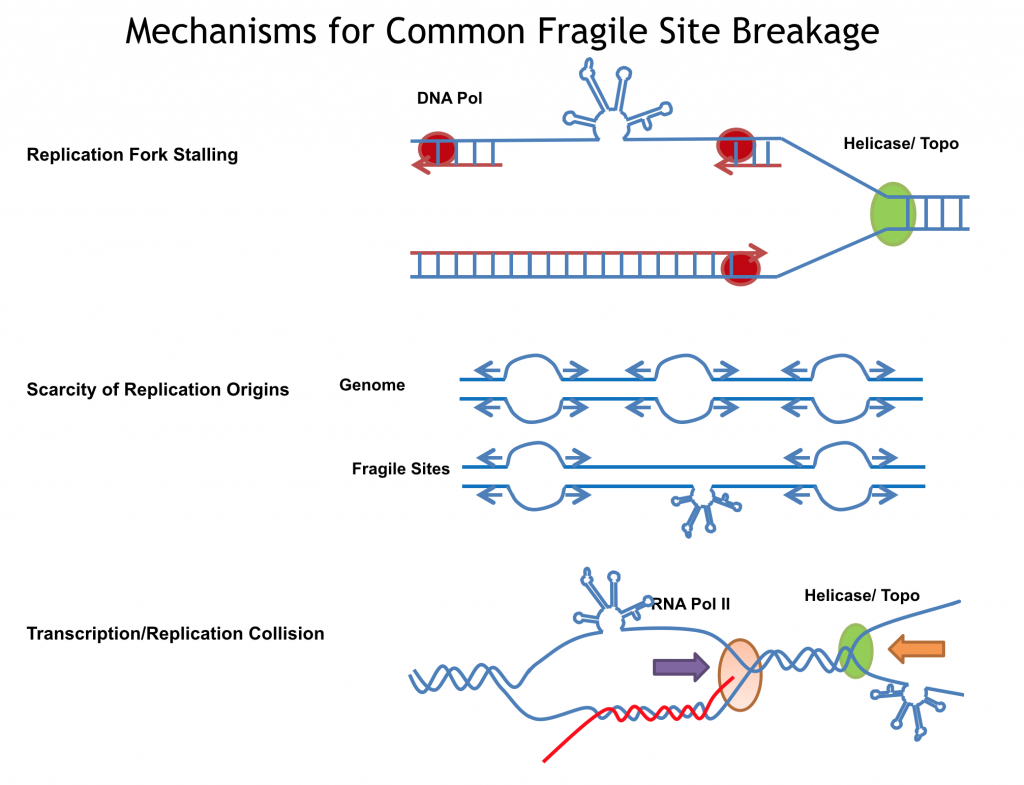
Both physiological DNA breaks occurring during DNA metabolic processes and pathological DNA breaks in response to a wide range of stresses, contribute to the outcome of human genome instability. DNA fragility generated by alternative DNA secondary structures is a known cause of many human diseases, and also occurs in normal DNA processes. Formation of alternative DNA secondary structures can arise from single-stranded DNA when the DNA duplex is unwound during DNA processes such as replication and transcription, and thus can be affected by cellular activities, nucleotide sequences, and chemical exposures. Current projects in the lab are to elucidate the mechanistic and functional features of DNA structure-driven fragility, and to understand the etiology of various rearrangement-driven cancers and repeat-expansion driven neurodegenerative diseases.
Ongoing projects include
(I) Genome-wide DNA Secondary Structure Analysis to Investigate DNA Fragility and Refine the Molecular Features of DNA Fragile Sites.
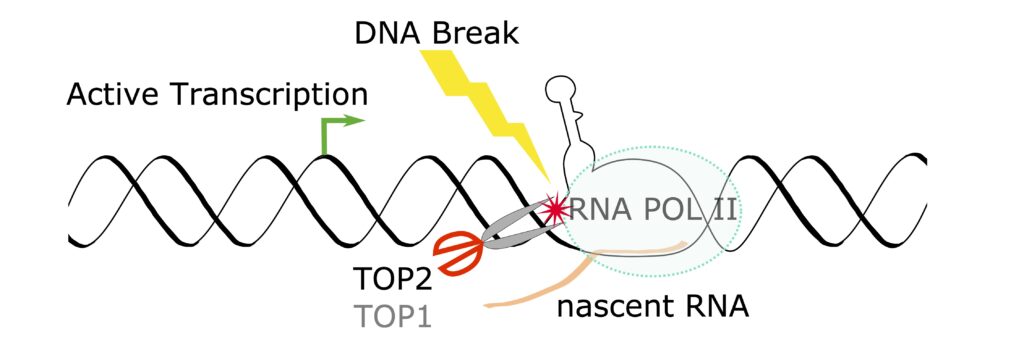
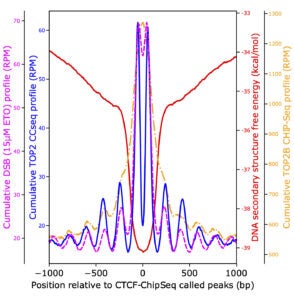 We for the first time carried out a computational evaluation of the entire available human genome sequence for optimal ability to fold single-stranded sequences into multiple stem-loop structures. With a list of regions capable of forming highly stable secondary structures, we discovered that RNA polymerase promoter-proximal pausing can be influenced by DNA secondary structures at pausing sites, and topoisomerase II directly contributes to the generation of double-stranded breaks at these sites. The secondary structure regions were also enriched at transcription start sites and CTCF-binding sites. The ultimate goal is to create a list of legitimate sites that are prone to DNA breakage caused by the secondary structure-forming mechanism(s), to evaluate genomic stress caused by endogenous and exogenous insults, and to facilitate future clinical use of fragile site breakage in disease diagnostics and screening.
We for the first time carried out a computational evaluation of the entire available human genome sequence for optimal ability to fold single-stranded sequences into multiple stem-loop structures. With a list of regions capable of forming highly stable secondary structures, we discovered that RNA polymerase promoter-proximal pausing can be influenced by DNA secondary structures at pausing sites, and topoisomerase II directly contributes to the generation of double-stranded breaks at these sites. The secondary structure regions were also enriched at transcription start sites and CTCF-binding sites. The ultimate goal is to create a list of legitimate sites that are prone to DNA breakage caused by the secondary structure-forming mechanism(s), to evaluate genomic stress caused by endogenous and exogenous insults, and to facilitate future clinical use of fragile site breakage in disease diagnostics and screening.
Related Publications: (1), (2), (3), (4), (5)
(II) Development of a DNA Fragility-based Test for Susceptibility to Gene Rearrangement-driven Cancers.
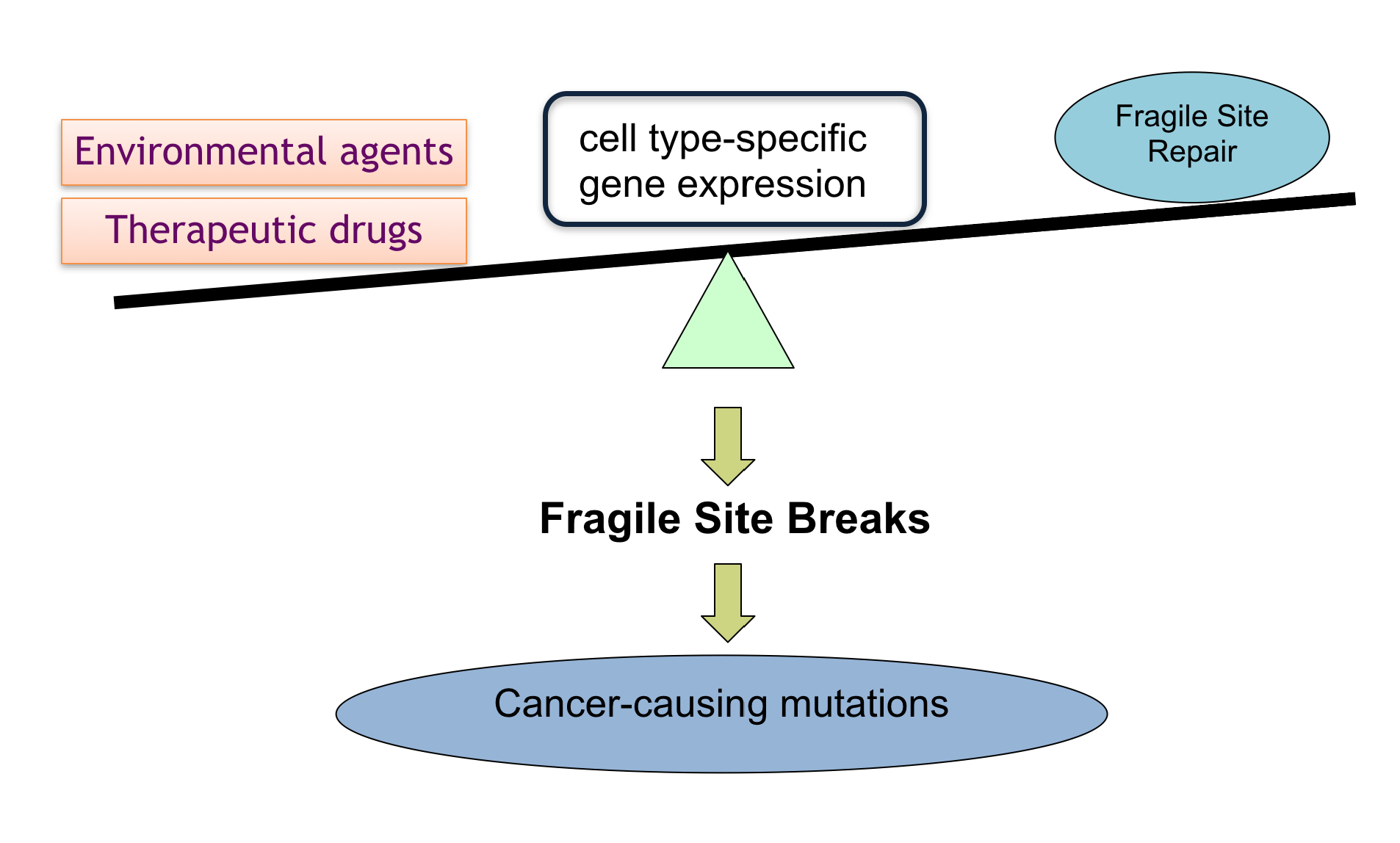
The DNA fragility test to detect DNA breakage – before cancer-causing rearrangements occur – offers an early indication of susceptibility to cancers, and facilitates prompt prevention and timely treatments. The current project is to develop a DNA test based on preferential breakage properties of rearrangement-participating genes. While this project currently focuses on the genes of papillary thyroid carcinoma and acute myeloid leukemia, this test could potentially be extended to other cancers, ultimately improving health through monitoring and early detection in at risk populations. This test will be particularly important for patients about to undergo chemotherapy. Identifying those at high risk for breakage, and thus rearrangement, before chemotherapy begins would allow clinicians to select non-DNA-breaking agents for treatment.
Related Publications: (1), (2), (3)
(III) Alternative DNA/RNA Secondary Structure in Genetic Diseases.
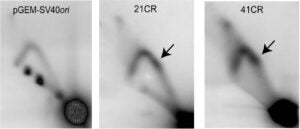
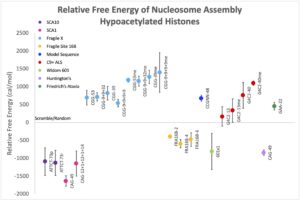
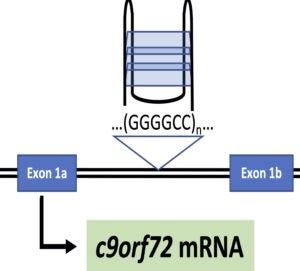
Over 30 human diseases, mostly neurological and muscular, are associated with expansion of repetitive DNA sequences. Repetitive DNA sequences have the ability to form a variety of secondary structures during normal cellular processes such as replication and transcription due to the unwinding of duplex DNA. To investigate the mechanism for repeat expansion and disease pathogenesis, we are examining nucleosome dynamics on repetitive DNA sequences and G-quadruplex secondary structure in C9orf72 repeat expansion driven ALS.