Research
Rewiring Organelle Proteostasis in Dormant Cancer Cells Leading to Metastasis
A small subpopulation of tumor cells can evade the immune response by entering a dormant state and developing resistance to chemotherapy drugs. This ability allows these cells to persist beyond the course of treatment, ultimately leading to cancer resurgence.
Metastatic relapse in cancer patients can occur within 2 years after treatment, but in some cases, it may take between 5 to 15 years for cancer to reappear. Disseminated Cancer Cells (DCCs) are often overlooked during cancer treatment since they are not detected by cell proliferation markers typically used in diagnostic settings. According to the World Health Organization (WHO), recurrent cancer accounts for a significant proportion of cancer-related deaths worldwide. Consequently, interfering with the mechanisms governing cell dormancy can have important ramifications for the development of new cancer therapies.
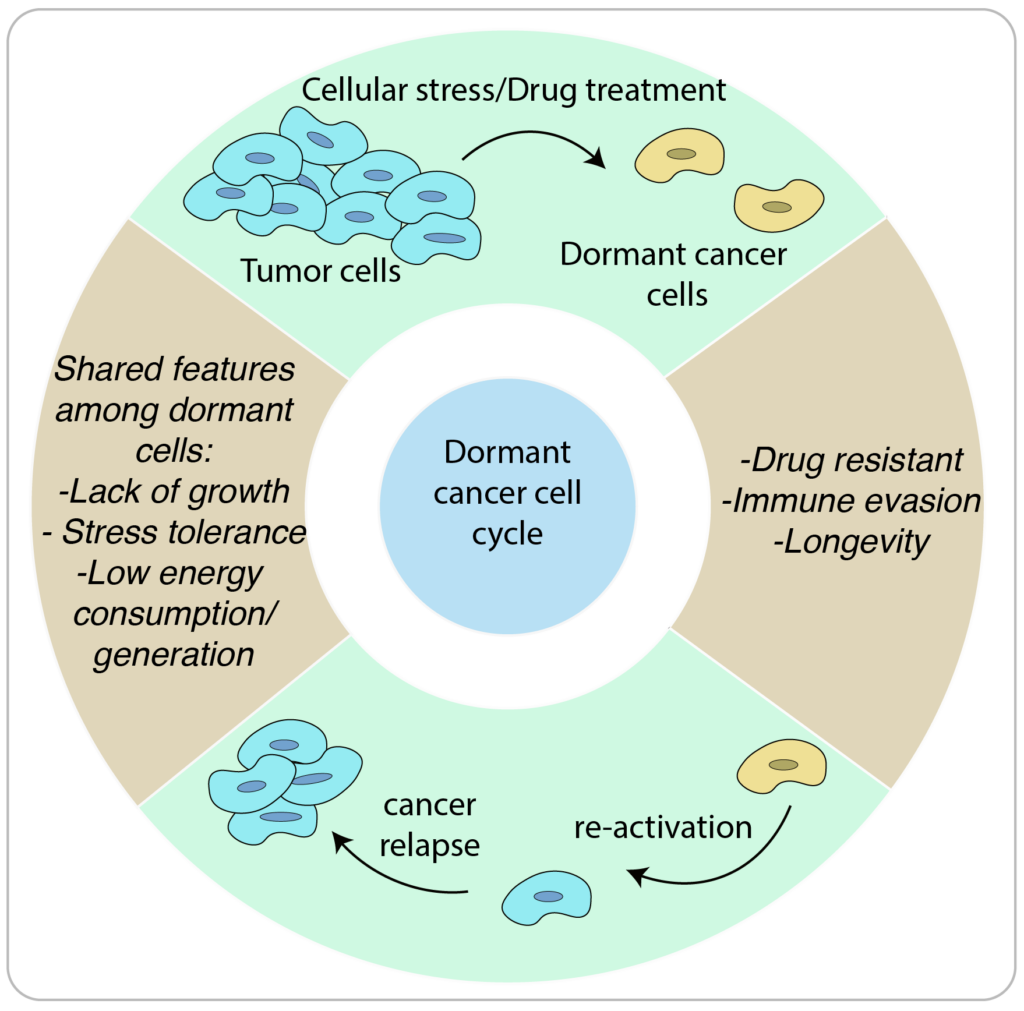
Dormancy is a quiescent state present in all kingdoms of life, encompassing plants, bacteria, fungi, and even humans. In this state, a cell ceases to divide and remains inactive until reactivated. This process enhances stress tolerance and longevity, providing the cell with the ability to persist during challenging conditions. Our laboratory is particularly focused on unraveling the regulatory mechanisms that govern the entry into and exit from the state of dormancy.
Our long-term objective is to uncover how dormant cells leverage their cellular machinery to endure and persist during stress. In the context of cancer, we aim to comprehend how these cells manage to evade the immune response and persist during drug treatments.
Currently, we employ a dormancy-inducible model to investigate the interconnection between protein synthesis and mitochondrial function under conditions of nutrient stress. This research will contribute to a deeper understanding of the molecular processes that underlie cellular dormancy and its implications for cancer survival and treatment resistance.
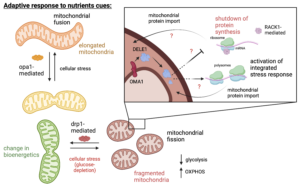
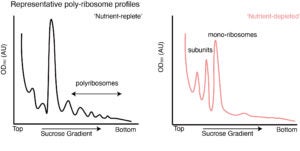
Under nutrient-limiting conditions, protein synthesis is down-regulated by a set of various effectors that bind to the active sites of ribosomes. Our research focuses on unraveling the regulatory mechanisms governing this process during the early stages of entry into and exit from the dormancy state.
Specifically, we aim to understand the intricate details of how these effectors modulate protein synthesis, contributing to the cell’s ability to adapt to nutrient stress. By investigating the regulatory events occurring during the early phases of dormancy, we hope to gain insights into the molecular pathways that orchestrate these processes.
This knowledge will not only enhance our understanding of cellular dormancy but also provide valuable information for developing strategies to manipulate these regulatory mechanisms. Such insights could have implications for various fields, including cancer research, where understanding and potentially controlling dormancy-related processes may offer new avenues for therapeutic interventions.
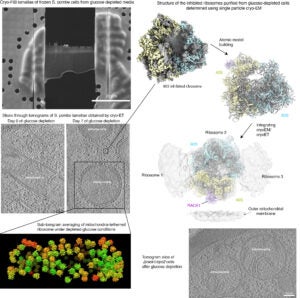
In our research, we employ a combination of protein biochemistry and structural biology approaches, incorporating high-resolution structure determination through cryo-electron microscopy. Additionally, we have established a collaboration with Dr. Simone Mattei’s group at EMBL Heidelberg. Together, we utilize cellular cryo-electron tomography and cryo-FIB milling techniques to explore dormant yeast cells, essentially capturing them in a frozen state.
This multi-faceted approach allows us to delve into the intricate details of cellular components and their interactions during dormancy. The use of cryo-electron microscopy provides high-resolution structural insights into the proteins involved, while cellular cryo-electron tomography and cryo-FIB milling enable us to observe the three-dimensional organization of dormant yeast cells at a cellular level.
By combining these cutting-edge techniques, we aim to uncover the structural and functional aspects of cellular components involved in dormancy. This collaborative effort enhances the comprehensiveness of our research, contributing valuable information to the broader understanding of dormancy-related processes in various biological contexts.
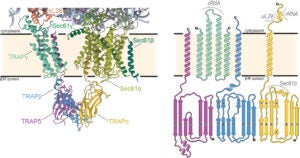
Protein Biogenesis at the Endoplasmic Reticulum and Mitochondria in Neurodegenerative Disease
Protein localization to subcellular compartments is a fundamental process essential for maintaining cell and organelle proteostasis. The decision to sort and target nascent proteins is made during synthesis on ribosomes and is encoded within the protein’s primary sequence as molecular “zip codes”. The focus of my lab’s research program is on elucidating the processes that govern the localization and targeting of nascent proteins to organelles, with specific attention directed towards the endoplasmic reticulum (ER) and mitochondria.

Dysfunction in these pathways result in protein aggregation, potentially leading to the onset of several neurodegenerative diseases. In particular, mutations in ER protein translocation and processing complexes have been linked to Congenital Disorders of Glycosylation (CDG). Additionally, impaired mitochondrial protein import has been associated with both Alzheimer’s and Parkinson’s diseases, underscoring the clinical relevance of our research program. Our research objectives are ambitiously set to: 1) Unravel the molecular intricacies of nascent chain channeling across ER chaperone and processing machineries during ER protein biogenesis. 2) Decode the mechanisms of mitochondrial protein sorting in the cytoplasm and the complex interplay between the sorting factors, followed by the receptors on the mitochondrial membrane.