All Publications
David Wissel, Madison M Mehlferber, Khue M Nguyen, Vasilii Pavelko, Elizabeth Tseng, Mark D Robinson, Gloria M Sheynkman. 2025. bioRXiv.

Luke Lambourne, Kaia Mattioli, Clarissa Santoso, Gloria Sheynkman, Sachi Inukai, Babita Kaundal, Anna Berenson, Kerstin Spirohn-Fitzgerald, Anukana Bhattacharjee, Elisabeth Rothman, Shaleen Shrestha, Florent Laval, Brent S Carroll, Stephen P Plassmeyer, Ryan J Emenecker, Zhipeng Yang, Deepa Bisht, Jared A Sewell, Guangyuan Li, Anisa Prasad, Sabrina Phanor, Ryan Lane, Devlin C Moyer, Toby Hunt, Dawit Balcha, Marinella Gebbia, Jean-Claude Twizere, Tong Hao, Alex S Holehouse, Adam Frankish, Josh A Riback, Nathan Salomonis, Michael A Cald. 2025. Molecular Cell.

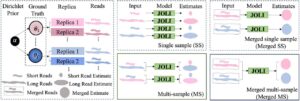
Multi-sample, multi-platform isoform quantification using empirical Bayes
Arghamitra Talukder, Shree Thavarekere, Madison Mehlferber, Gloria M Sheynkman, David A Knowles. 2025. bioRxiv.
Aabida Saferali, Anastacia N. Wienecke, Zhonghui Xu, Tao Liu, Gloria M. Sheynkman, Craig P. Hersh, Michael H. Cho, Edwin K. Silverman, Xiaobo Zhou, Carole L. Wilson, Lynn M. Schnapp, Scott H. Randell, Silvia B. V. Ramos, Alain Laederach, Christopher Vollmers, Peter J. Castaldi. 2025. European Respiratory Journal.
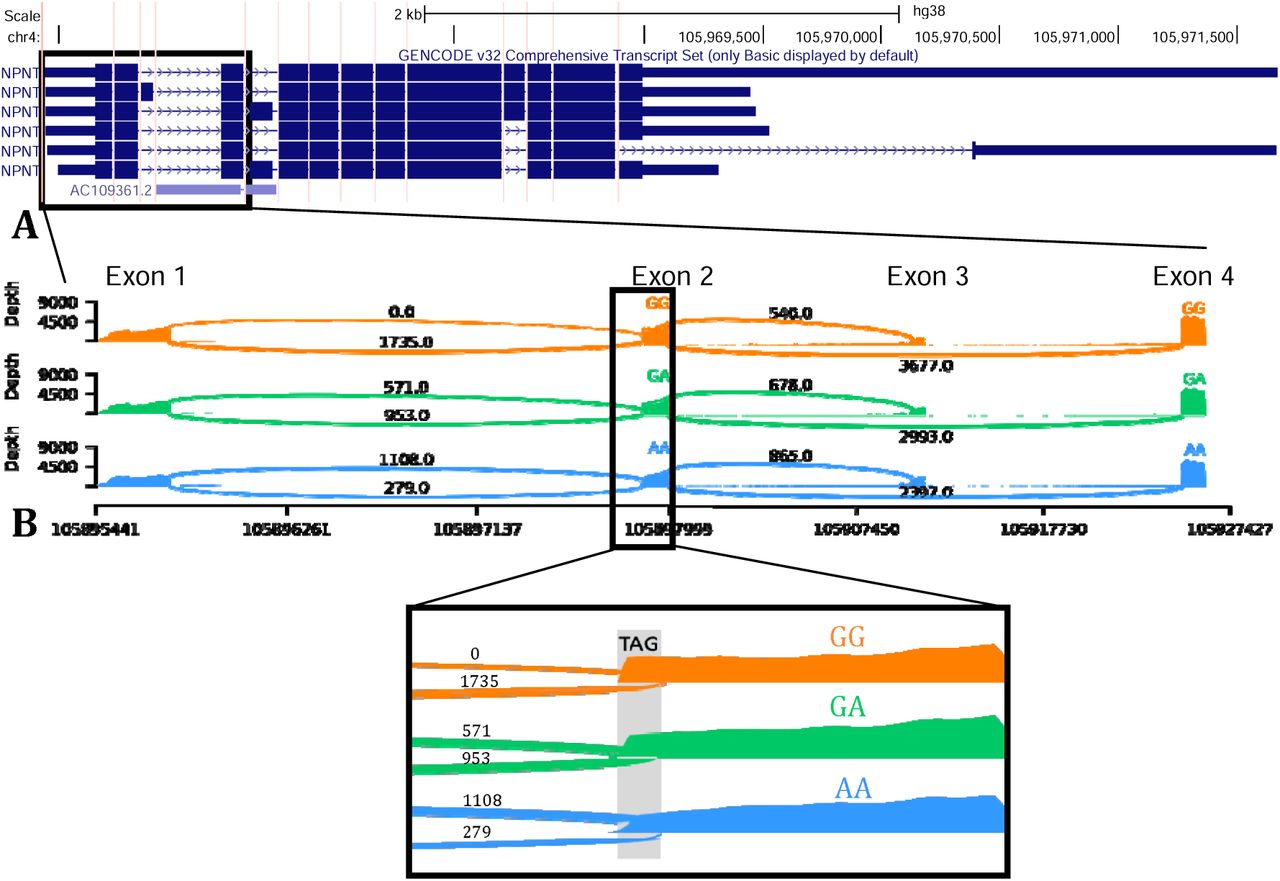
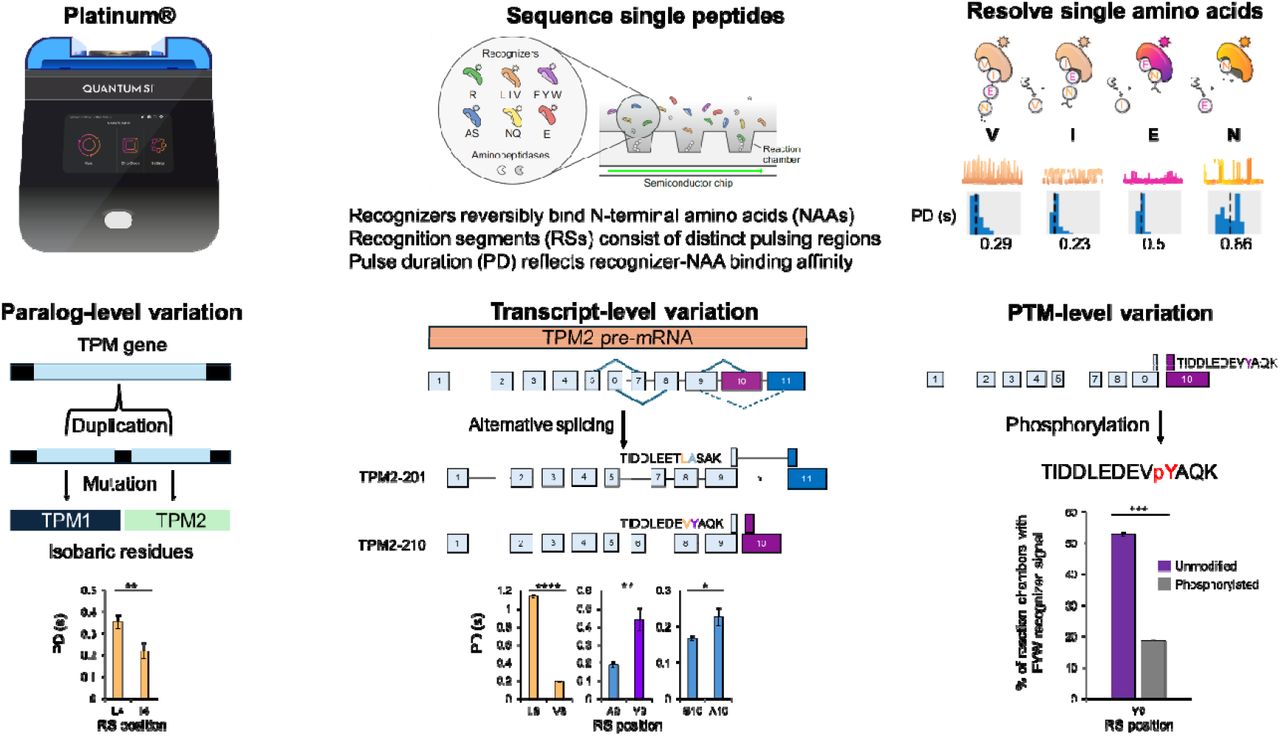
Natchanon Sittipongpittaya, Kenneth A. Skinner, Erin D. Jeffery, Emily F. Watts, Gloria M. Sheynkman. 2024. bioRxiv
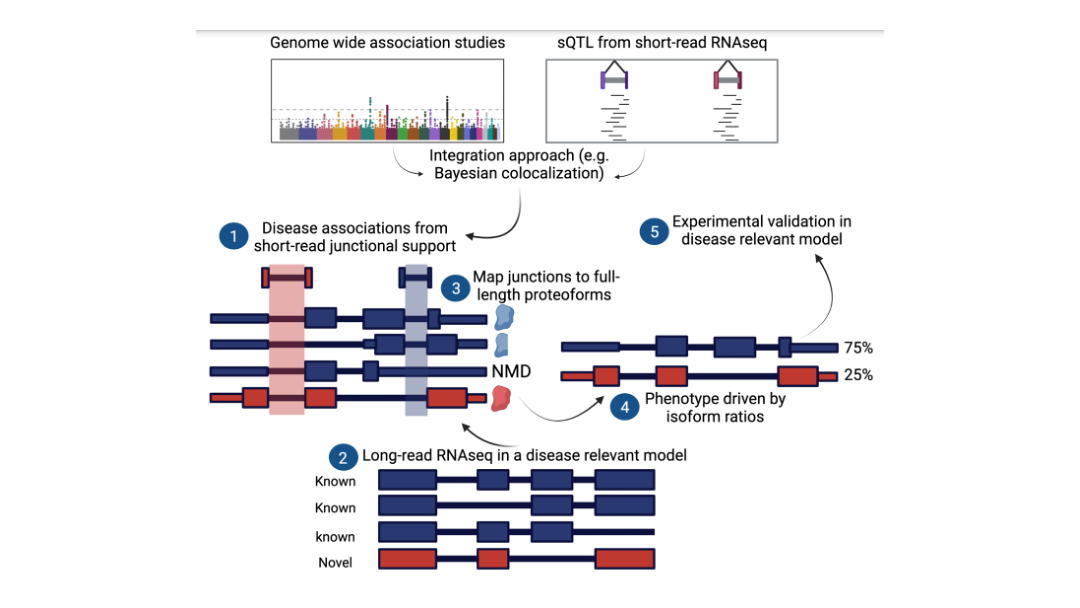
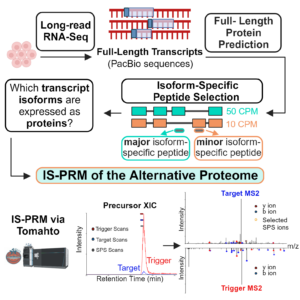
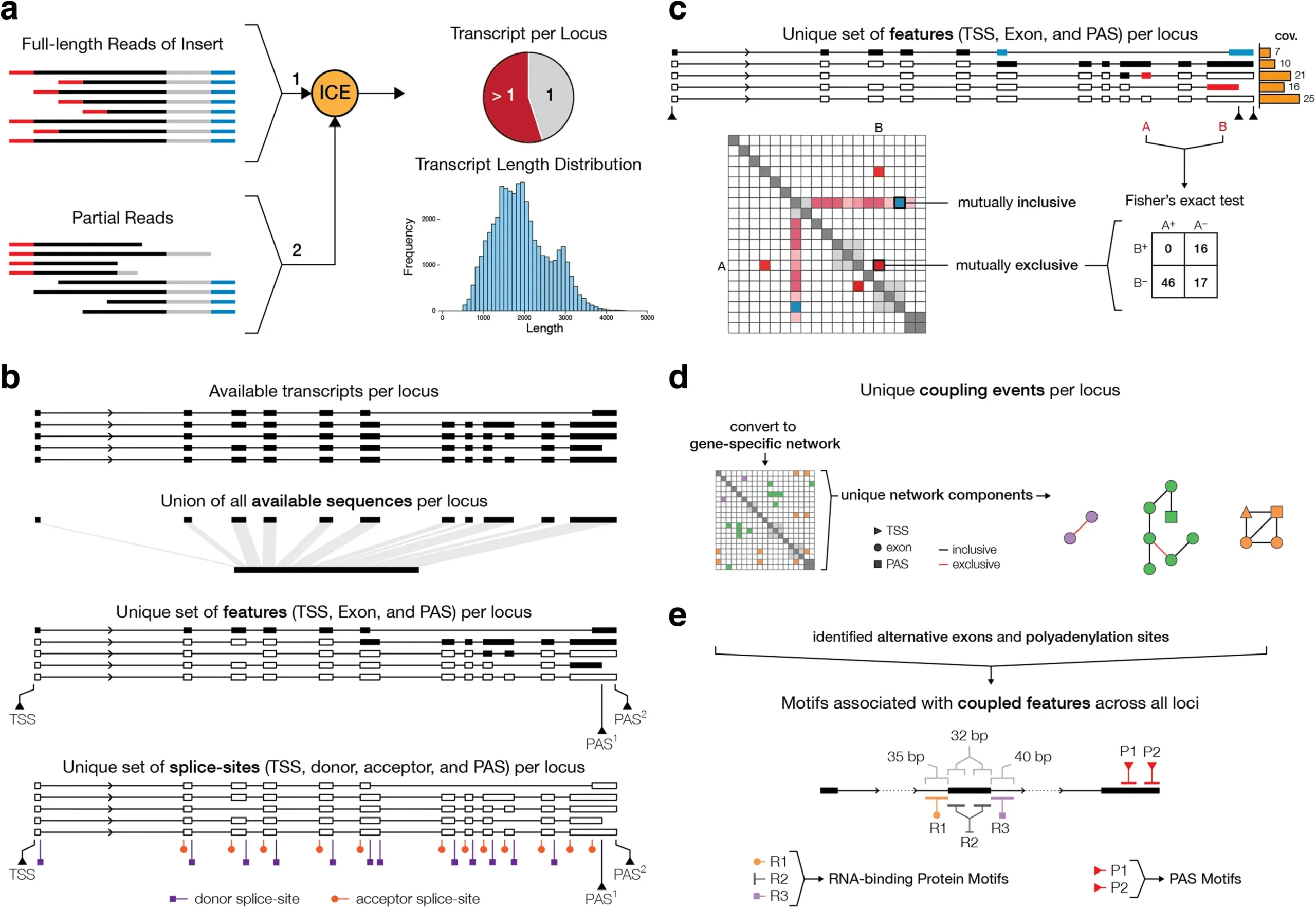
Long-read proteogenomics to connect disease-associated sQTLs to 2 the protein isoform effectors of disease
Abdullah Abood, Larry D. Mesner, Erin D. Jeffery , Mayank Murali , Micah D. Lehe , Jamie Saquing , Charles R. Farber, Gloria M. Sheynkman. The American Journal of Human Genetics. 2024. 111(9): 1914-1931.
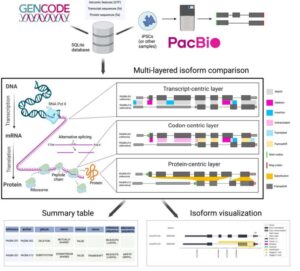
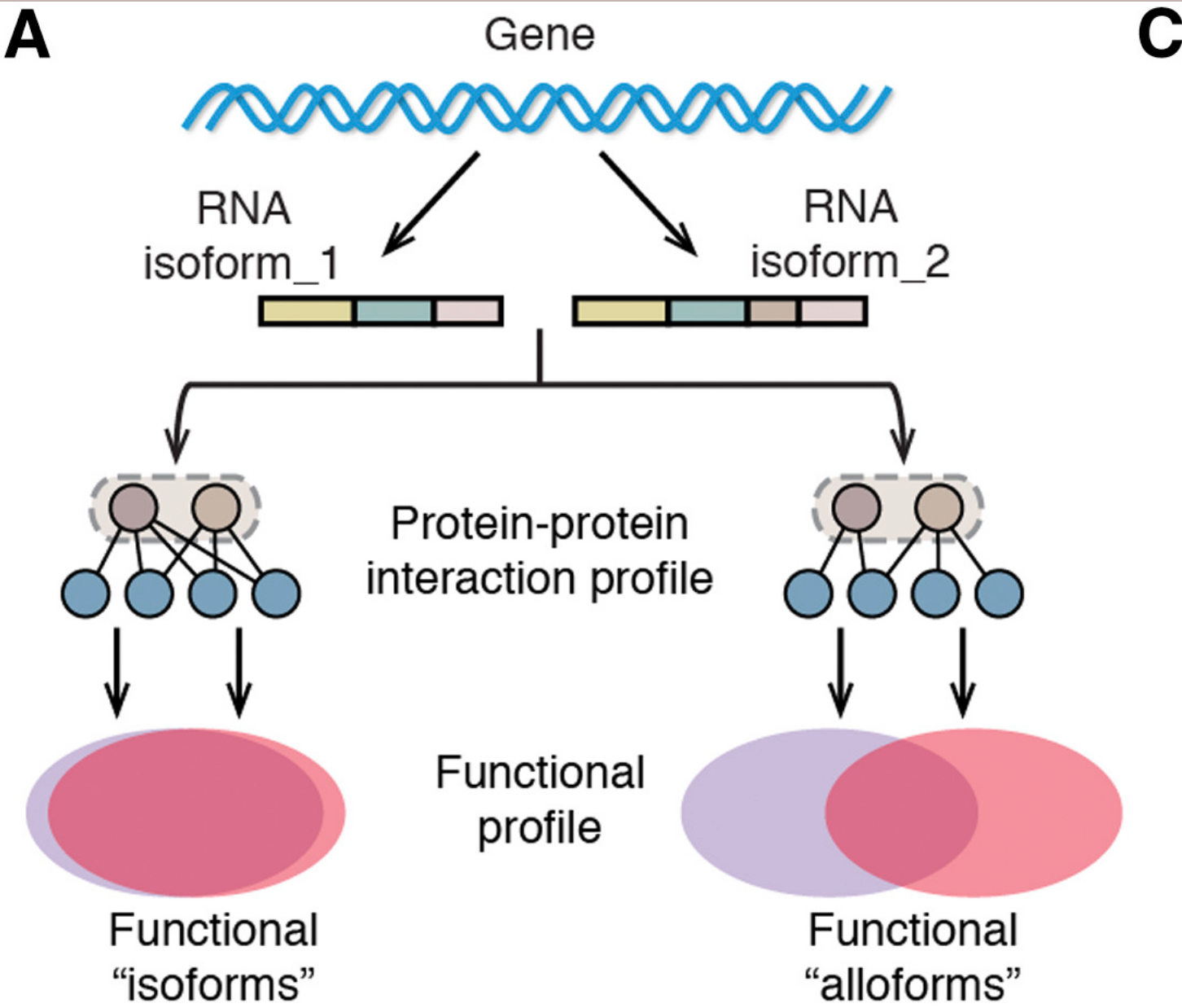
Biosurfer for systematic tracking of regulatory mechanisms leading to protein isoform diversity
Murali M, Saquing J, Lu S, Gao Z, Jordan B, Wakefield ZP, Fiszbein A, Cooper DR, Castaldi PJ, Korkin D, Sheynkman G. Biosurfer for systematic tracking of regulatory mechanisms leading to protein isoform diversity. bioRxiv [Preprint]. 2024 Mar
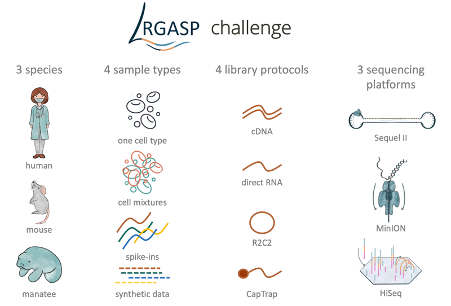
Systematic assessment of long-read RNA-seq methods for transcript identification and quantification
F.J. Pardo-Palacios, D. Wang, F. Reese, M. Diekhans, S. Carbonell-Sala, B. Williams, J.E. Loveland, M. De María, M.S. Adams, G. Balderrama-Gutierrez, A.K. Behera, J.M. Gonzalez, T. Hunt, J. Lagarde, C.E. Liang, H. Li, M. Jerryd Meade, D.A. Moraga Amador, A.D. Prjibelski, I. Birol, H. Bostan, A.M. Brooks, M. Hasan Çelik, Y. Chen, M.R.M. Du, C. Felton, J. Göke, S. Hafezqorani, R. Herwig, H. Kawaji, J. Lee, J. Liang Li, M. Lienhard, A. Mikheenko, D. Mulligan, K. Ming Nip, M. Pertea, M.E. Ritchie, A.D. Sim, A.D. Tang, Y. Kei Wan, C. Wang, B.Y. Wong, C. Yang, I. Barnes, A. Berry, S. Capella, N. Dhillon, J.M. Fernandez-Gonzalez, L. Ferrández-Peral, N. Garcia-Reyero, S. Goetz, C. Hernández-Ferrer, L. Kondratova, T. Liu, A. Martinez-Martin, C. Menor, J. Mestre-Tomás, J.M. Mudge, N.G. Panayotova, A. Paniagua, D. Repchevsky, E. Rouchka, B. Saint-John, E. Sapena, Leon Sheynkman, M. Laird Smith, M.M. Suner, H. Takahashi, I.A. Youngworth, P. Carninci, N.D. Denslow, R. Guigó, M.E. Hunter, H.U. Tilgner, B.J. Wold, C. Vollmers, A. Frankish, K. Fai Au, Gloria M. Sheynkman, A. Mortazavi, A. Conesa, A.N. Brooks. Nature Methods. 2024. 21: 1349-1363.
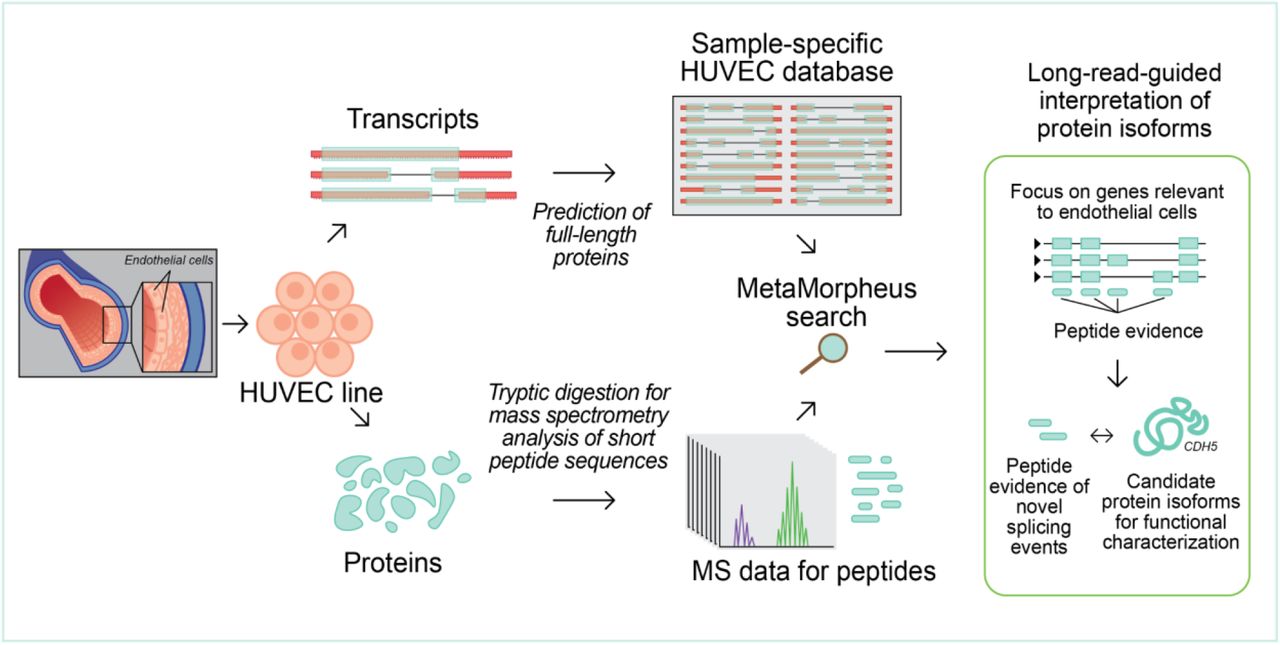
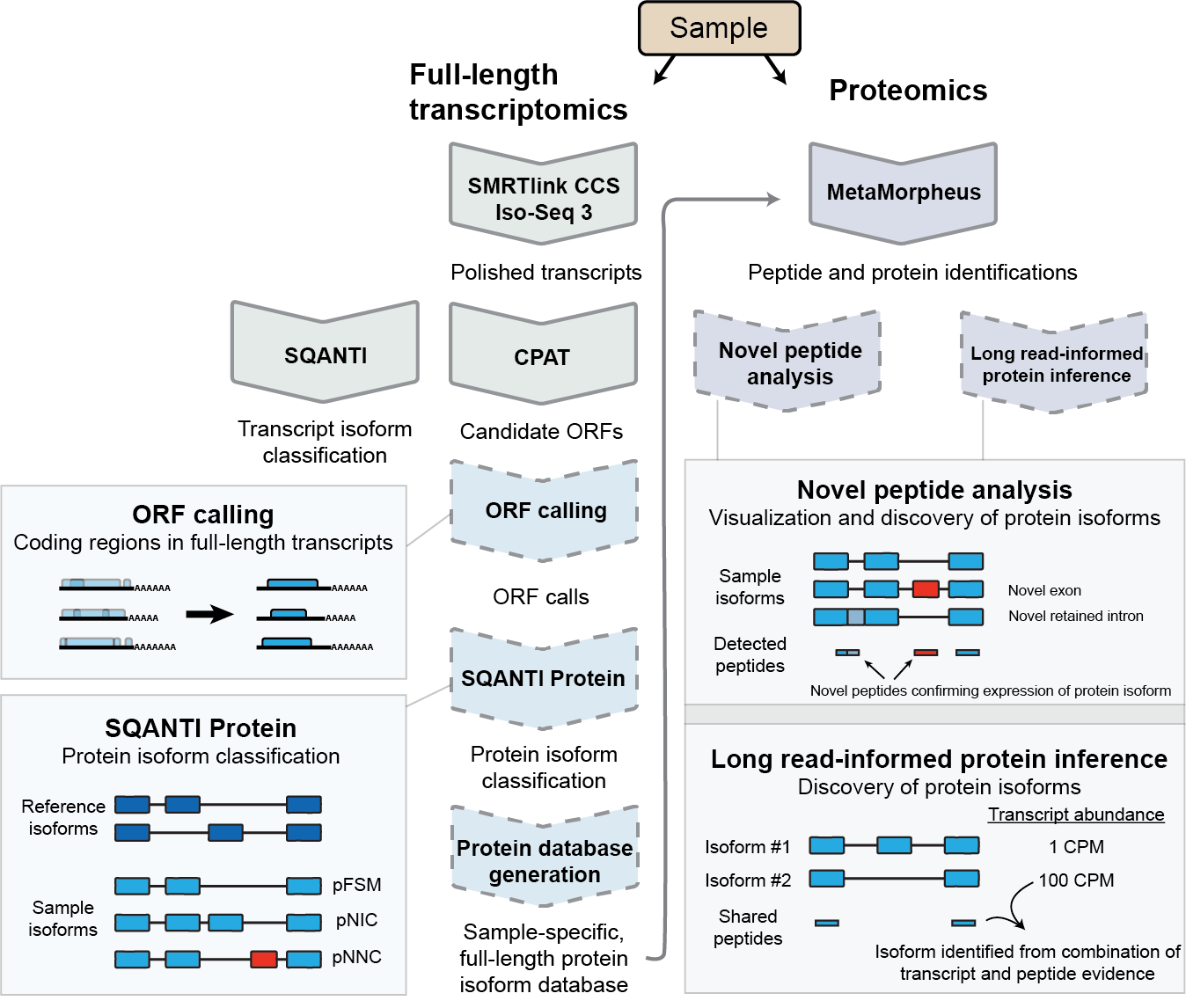
Enhanced protein isoform characterization through long-read proteogenomics
Rachel M. Miller, Ben T. Jordan, Madison M. Mehlferber, Erin D. Jeffery, Christina Chatzipantsiou, Simi Kaur, Robert J. Millikin, Yunxiang Dai, Simone Tiberi, Peter J. Castaldi, Michael R. Shortreed, Chance John Luckey, Ana Conesa, Lloyd M. Smith, Anne Deslattes Mays, Gloria M. Sheynkman. Genome Biology. 2022. 23(1): 69.
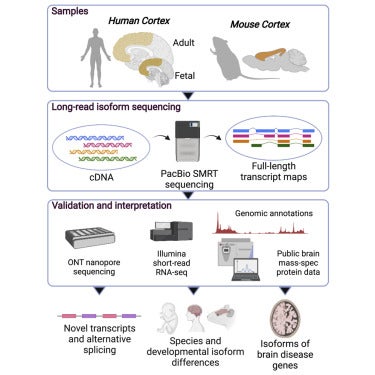
Full-length transcript sequencing of human and mouse cerebral cortex identifies widespread isoform diversity and alternative splicing
Szi Kay Leung, Aaron R. Jeffries, Isabel Castanho, Ben T. Jordan, Karen Moore, Jonathan P. Davies, Emma L. Dempster, Nicholas J. Bray, Paul O’Neill, Elizabeth Tseng, Zeshan Ahmed, David A. Collier, Erin D. Jeffery, Shyam Prabhakar, Leonard Schalkwyk, Connor Jops, Michael J. Gandal, Gloria M. Sheynkman, Elis Hannon, Jonathan Mill. Cell Reports. 2021. 37(7): 110022.
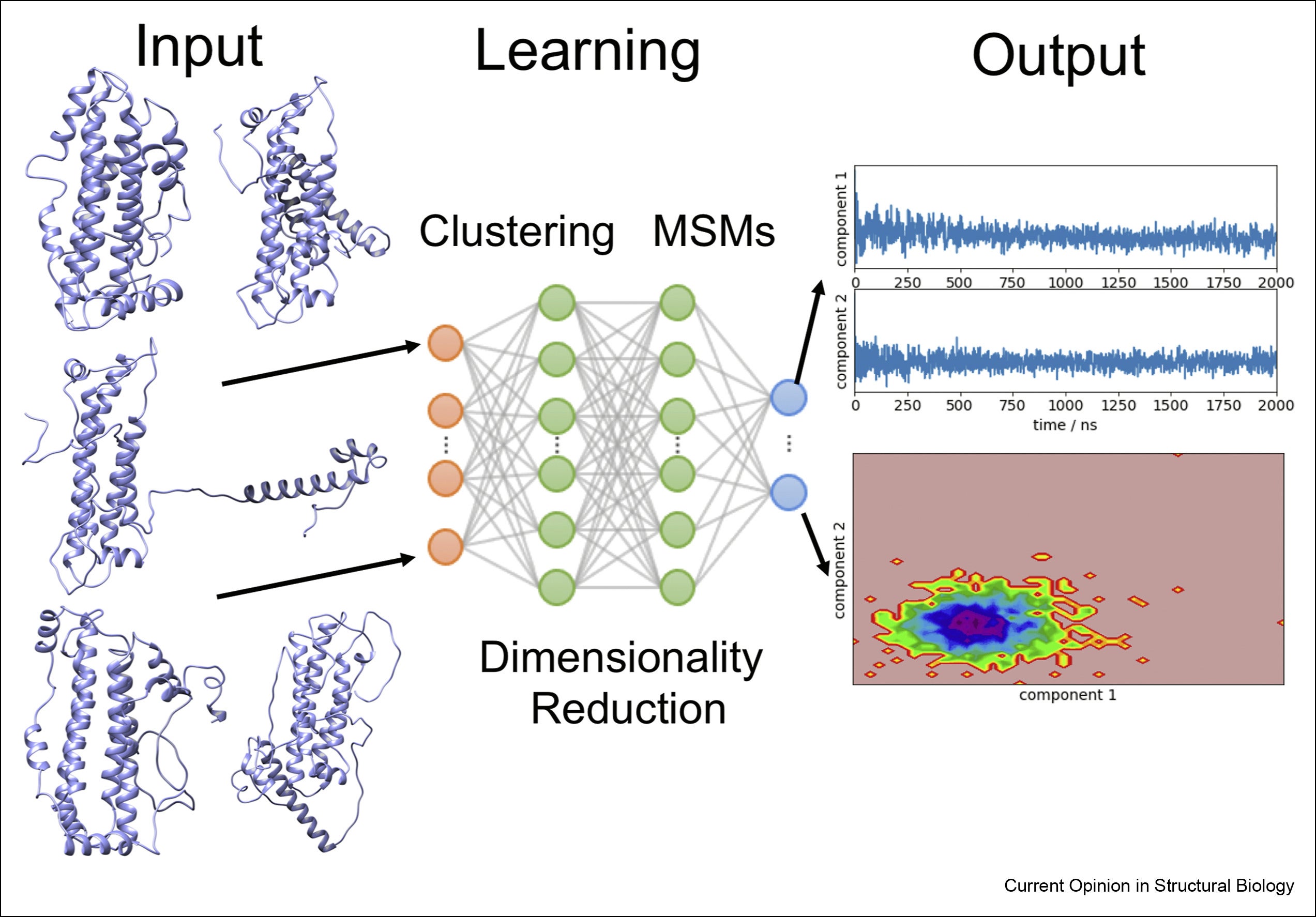
Interpretable artificial intelligence and exascale molecular dynamics simulations to reveal kinetics: Applications to Alzheimer’s disease
William Martin, Gloria Sheynkman, Felice C. Lightstone, Ruth Nussinov, Feixiong Cheng. Current Opinion in Structural Biology. 2022. 72: 103-113.
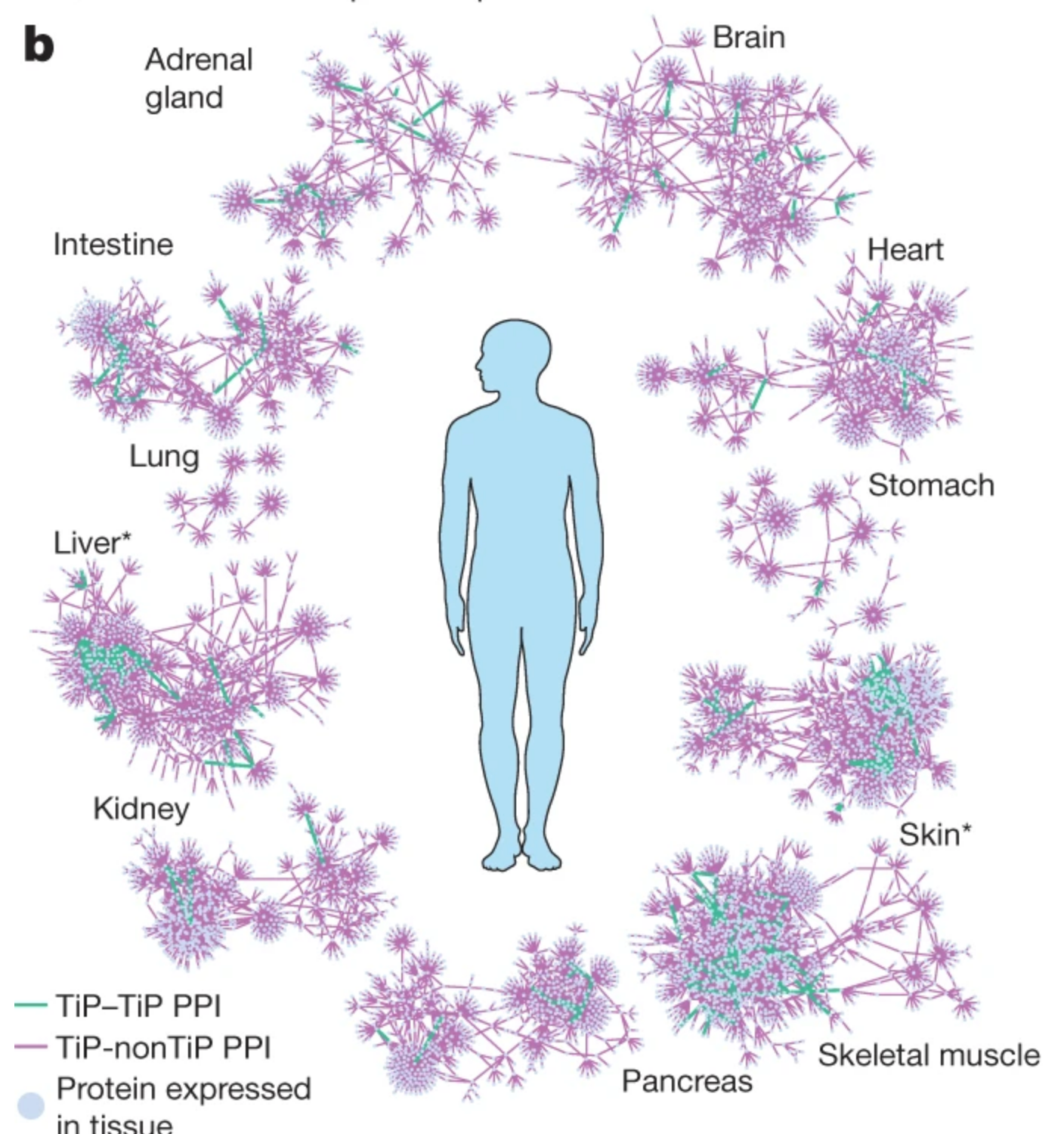
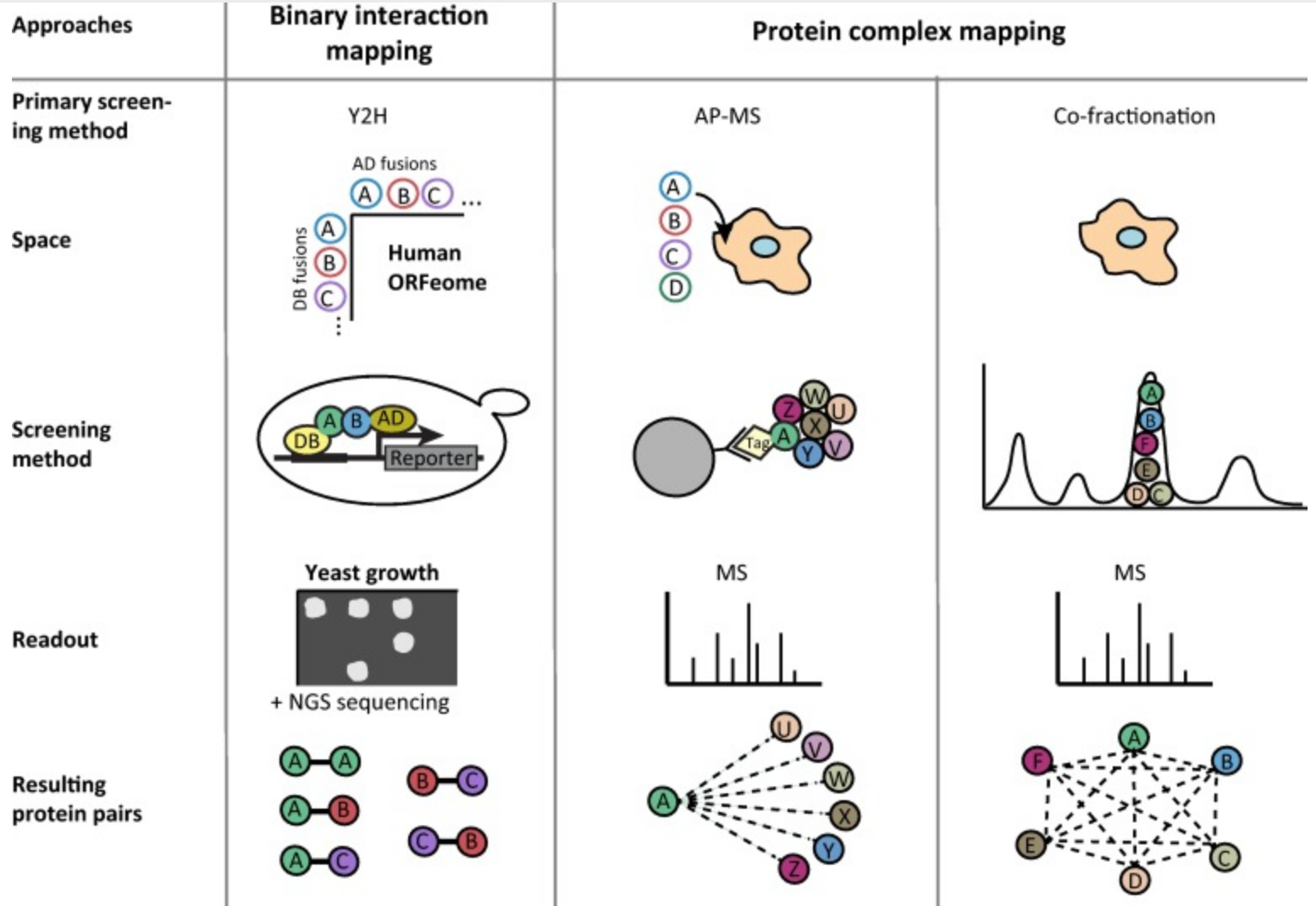
A reference map of the human binary protein interactome
Katja Luck, Dae-Kyum Kim, Luke Lambourne, Kerstin Spirohn, Bridget E. Begg, Wenting Bian, Ruth Brignall, Tiziana Cafarelli, Francisco J. Campos-Laborie, Benoit Charloteaux, Dongsic Choi, Atina G. Coté, Meaghan Daley, Steven Deimling, Alice Desbuleux, Amélie Dricot, Marinella Gebbia, Madeleine F. Hardy, Nishka Kishore, Jennifer J. Knapp, István A. Kovács, Irma Lemmens, Miles W. Mee, Joseph C. Mellor, Carl Pollis, Carles Pons, Aaron D. Richardson, Sadie Schlabach, Bridget Teeking, Anupama Yadav, Mariana Babor, Dawit Balcha, Omer Basha, Christian Bowman-Colin, Suet-Feung Chin, Soon Gang Choi, Claudia Colabella, Georges Coppin, Cassandra D’Amata, David De Ridder, Steffi De Rouck, Miquel Duran-Frigola, Hanane Ennajdaoui, Florian Goebels, Liana Goehring, Anjali Gopal, Ghazal Haddad, Elodie Hatchi, Mohamed Helmy, Yves Jacob, Yoseph Kassa, Serena Landini, Roujia Li, Natascha van Lieshout, Andrew MacWilliams, Dylan Markey, Joseph N. Paulson, Sudharshan Rangarajan, John Rasla, Ashyad Rayhan, Thomas Rolland, Adriana San-Miguel, Yun Shen, Dayag Sheykhkarimli, Gloria M. Sheynkman, Eyal Simonovsky, Murat Taşan, Alexander Tejeda, Vincent Tropepe, Jean-Claude Twizere, Yang Wang, Robert J. Weatheritt, Jochen Weile, Yu Xia, Xinping Yang, Esti Yeger-Lotem, Quan Zhong, Patrick Aloy, Gary D. Bader, Javier De Las Rivas, Suzanne Gaudet, Tong Hao, Janusz Rak, Jan Tavernier, David E. Hill, Marc Vidal, Frederick P. Roth & Michael A. Calderwood. Nature. 2020. 580(7803): 402-408.
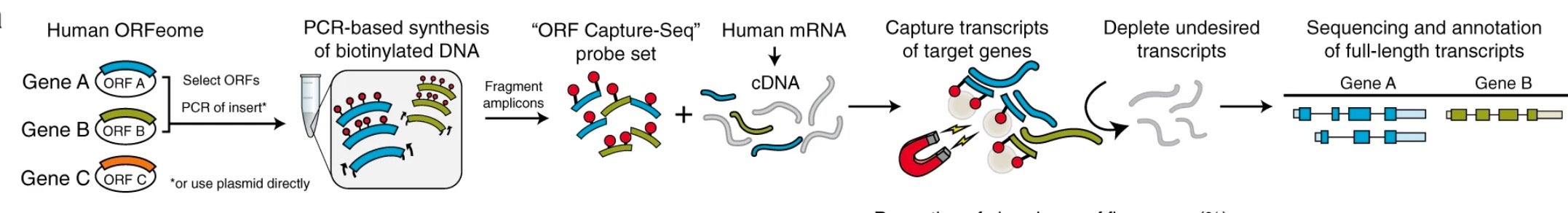
ORF Capture-Seq as a versatile method for targeted identification of full-length isoforms
Gloria M. Sheynkman, Katharine S. Tuttle, Florent Laval, Elizabeth Tseng, Jason G. Underwood, Liang Yu, Da Dong, Melissa L. Smith, Robert Sebra, Luc Willems, Tong Hao, Michael A. Calderwood, David E. Hill & Marc Vidal . 2020. Nature communications. 11(1): 2326.
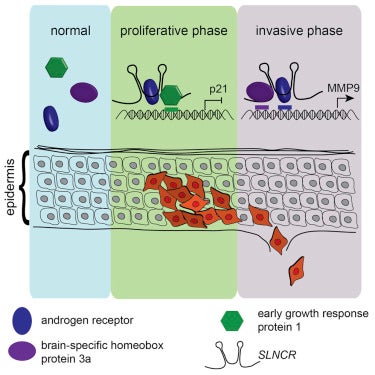
The lncRNA SLNCR Recruits the Androgen Receptor to EGR1-Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor
Karyn Schmidt, Johanna S. Carroll, Elaine Yee, Dolly D. Thomas, Leon Wert-Lamas, Steven C. Neier, Gloria Sheynkman, Justin Ritz, Carl D. Novina. 2019. Cell Reports. 27(8): 2493-2507.e4.
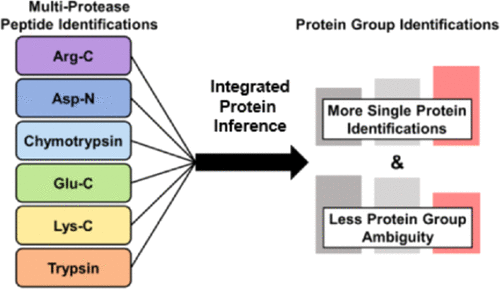
Improved Protein Inference from Multiple Protease Bottom-Up Mass Spectrometry Data
Rachel M. Miller, Robert J. Millikin, Connor V. Hoffmann, Stefan K. Solntsev, Gloria M. Sheynkman, Michael R. Shortreed, Lloyd M. Smith. 2019. Journal of Proteome Research. 18(9): 3429-3438.
Full-length mRNA sequencing uncovers a widespread coupling between transcription initiation and mRNA processing
Seyed Yahya Anvar, Guy Allard, Elizabeth Tseng, Gloria M. Sheynkman, Eleonora de Klerk, Martijn Vermaat, Raymund H. Yin, Hans E. Johansson, Yavuz Ariyurek, Johan T. den Dunnen, Stephen W. Turner & Peter A. C. ‘t Hoen. 2018. Genome biology. 19(1): 46.
Proteome-Scale Human Interactomics
Katja Luck, Gloria M. Sheynkman, Ivy Zhang, Marc Vidal. 2017. Trends in Biochemical Sciences. 42(5): 342-354.
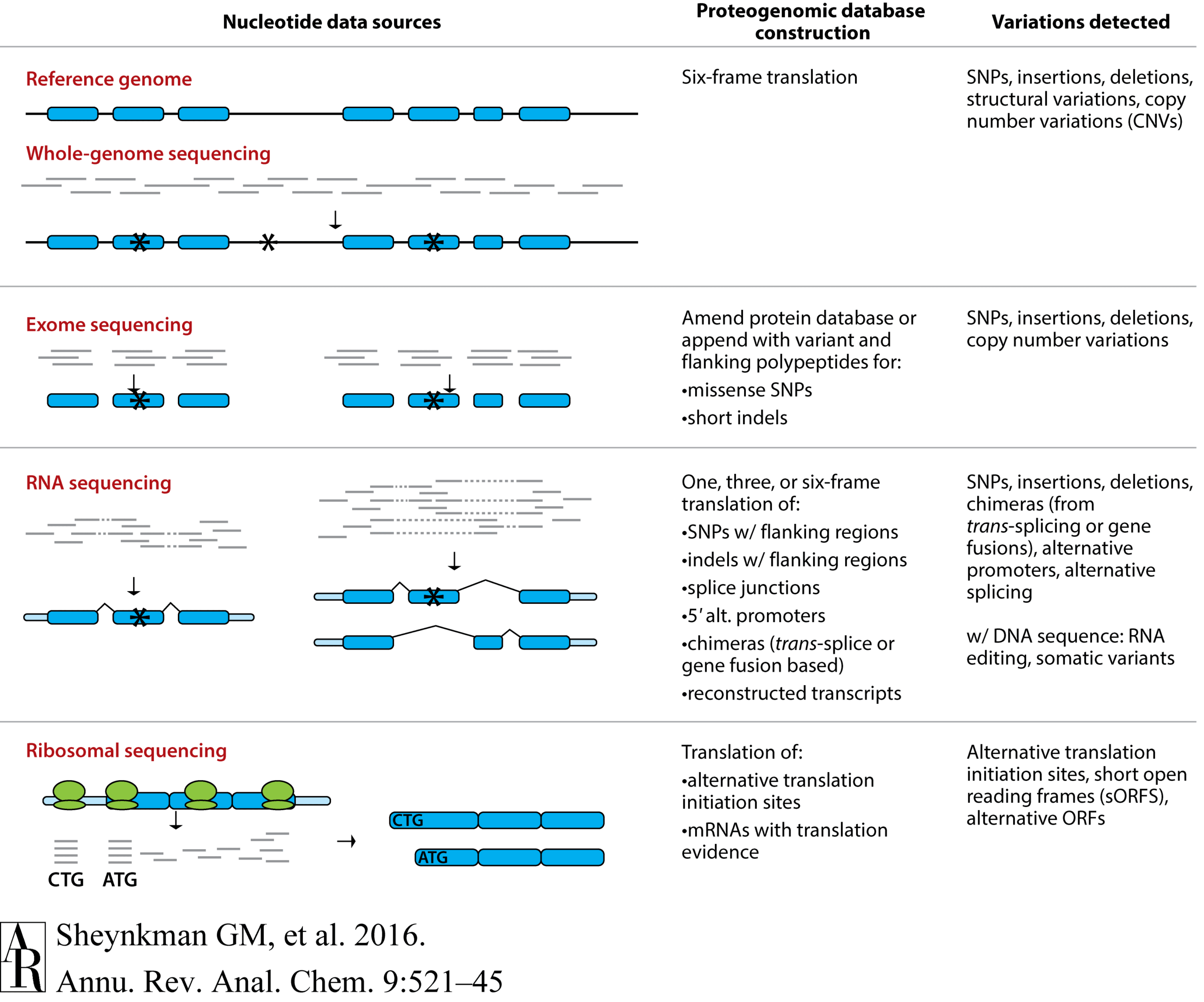
Proteogenomics: Integrating Next-Generation Sequencing and Mass Spectrometry to Characterize Human Proteomic Variation
Gloria M. Sheynkman, Michael R. Shortreed, Anthony J. Cesnik, and Lloyd M. Smith. 2016; Annual review of analytical chemistry (Palo Alto, Calif.). 9(1): 521-45.
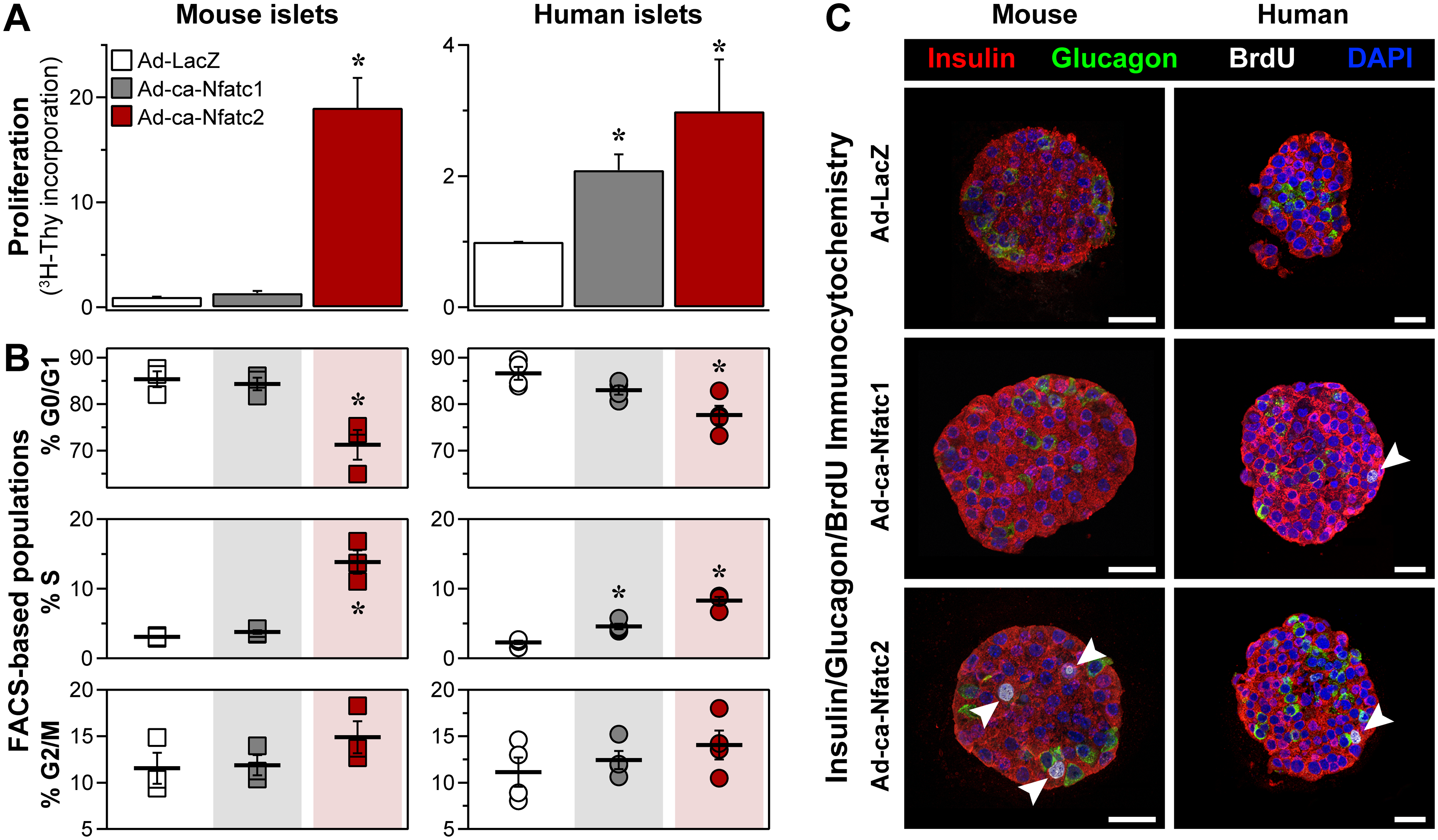
The Transcription Factor Nfatc2 Regulates β-Cell Proliferation and Genes Associated with Type 2 Diabetes in Mouse and Human Islets
Mark P. Keller, Pradyut K. Paul, Mary E. Rabaglia, Donnie S. Stapleton, Kathryn L. Schueler, Aimee Teo Broman, Shuyun Isabella Ye, Ning Leng, Christopher J. Brandon, Elias Chaibub Neto, Christopher L. Plaisier, Shane P. Simonett, Melkam A. Kebede, Gloria M. Sheynkman, Mark A. Klein, Nitin S. Baliga, Lloyd M. Smith, Karl W. Broman, Brian S. Yandell, Christina Kendziorski, Alan D. Attie. 2016. PLoS Genetics. 12(12): e1006466.
Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing
Xinping Yang, Jasmin Coulombe-Huntington, Shuli Kang, Gloria M. Sheynkman, Tong Hao, Aaron Richardson, Song Sun, Fan Yang, Yun A. Shen, Ryan R. Murray, Kerstin Spirohn, Bridget E. Begg, Miquel Duran-Frigola, Andrew MacWilliams, Samuel J. Pevzner, Quan Zhong, Shelly A. Wanamaker, Stanley Tam, Lila Ghamsari, Nidhi Sahni, Song Yi, Maria D. Rodriguez, Dawit Balcha, Guihong Tan, Michael Costanzo, Brenda Andrews, Charles Boone, Xianghong J. Zhou, Kourosh Salehi-Ashtiani, Benoit Charloteaux, Alyce A. Chen, Michael A. Calderwood, Patrick Aloy, Frederick P. Roth, David E. Hill, Lilia M. Iakoucheva, Yu Xia, Marc Vidal. 2016. Cell. 164(4): 805-17.
Global Identification of Protein Post-translational Modifications in a Single-Pass Database Search
Michael R. Shortreed, Craig D. Wenger, Brian L. Frey, Gloria M. Sheynkman, Mark Scalf, Mark P. Keller, Alan D. Attie, Lloyd M. Smith. 2015; Journal of Proteome Research. 14(11): 4714-20.
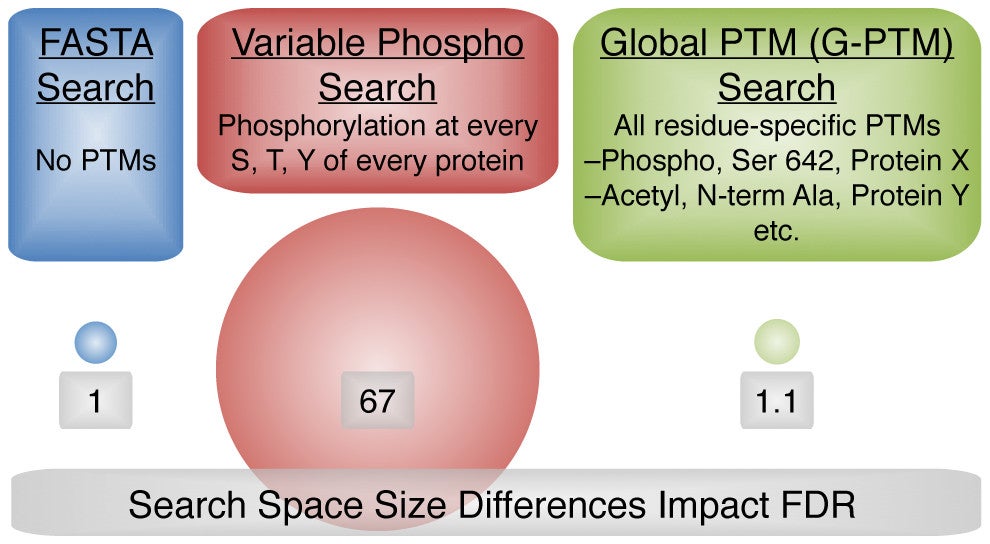
Human Proteomic Variation Revealed by Combining RNA-Seq Proteogenomics and Global Post-Translational Modification (G-PTM) Search Strategy
Anthony J. Cesnik, Michael R. Shortreed, Gloria M. Sheynkman, Brian L. Frey, Lloyd M. Smith. 2015. Journal of Proteome Research. 15(3): 800-8.
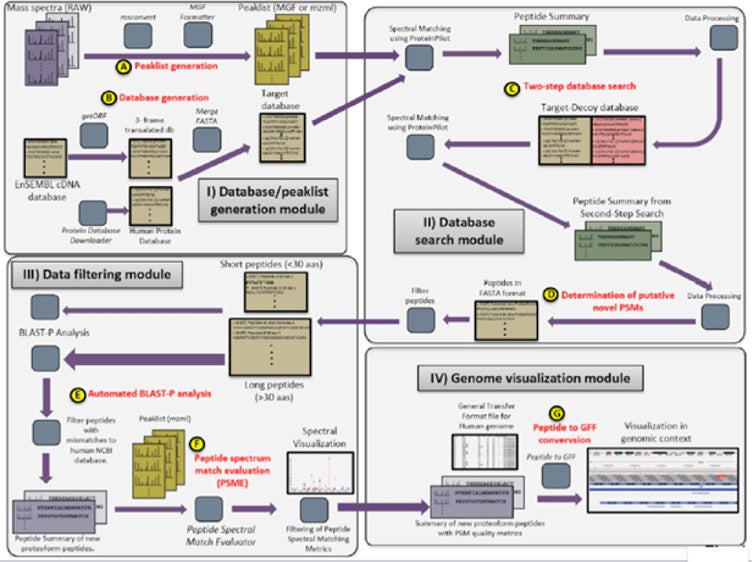
Flexible and accessible workflows for improved proteogenomic analysis using the Galaxy framework
Pratik D. Jagtap, James E. Johnson, Getiria Onsongo, Fredrik W. Sadler, Kevin Murray, Yuanbo Wang, Gloria M. Shenykman, Sricharan Bandhakavi, Lloyd M. Smith, Timothy J. Griffin. 2014. Journal of Proteome Research. 13(12): 5898-908.
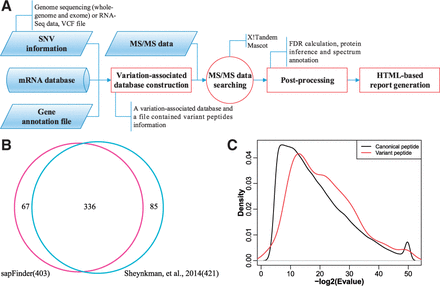
sapFinder: an R/Bioconductor package for detection of variant peptides in shotgun proteomics experiments
Bo Wen, Shaohang Xu, Gloria M. Sheynkman, Qiang Feng, Liang Lin, Quanhui Wang, Xun Xu, Jun Wang, Siqi Liu. 2014. Bioinformatics (Oxford, England). 30(21): 3136-8.
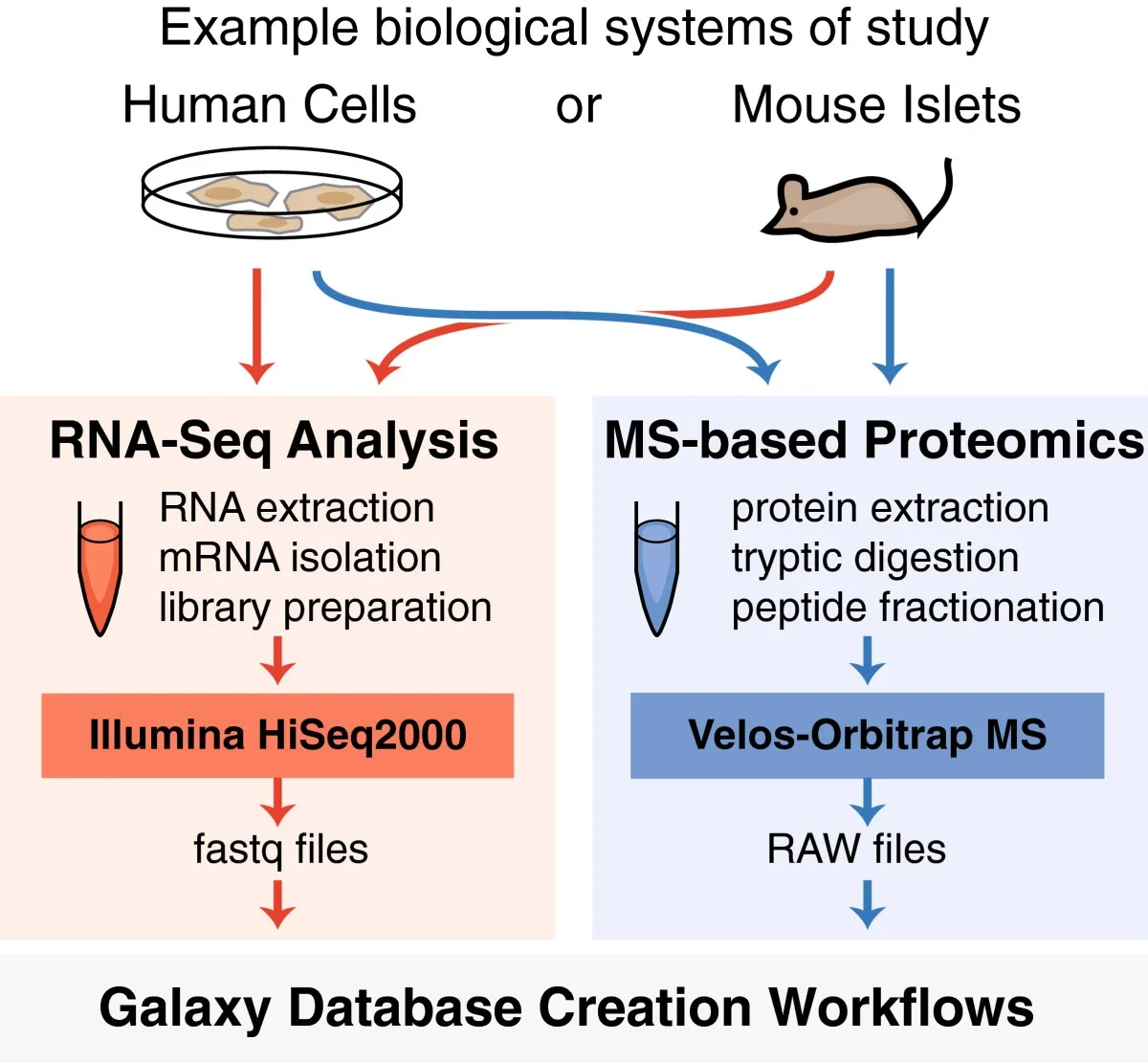
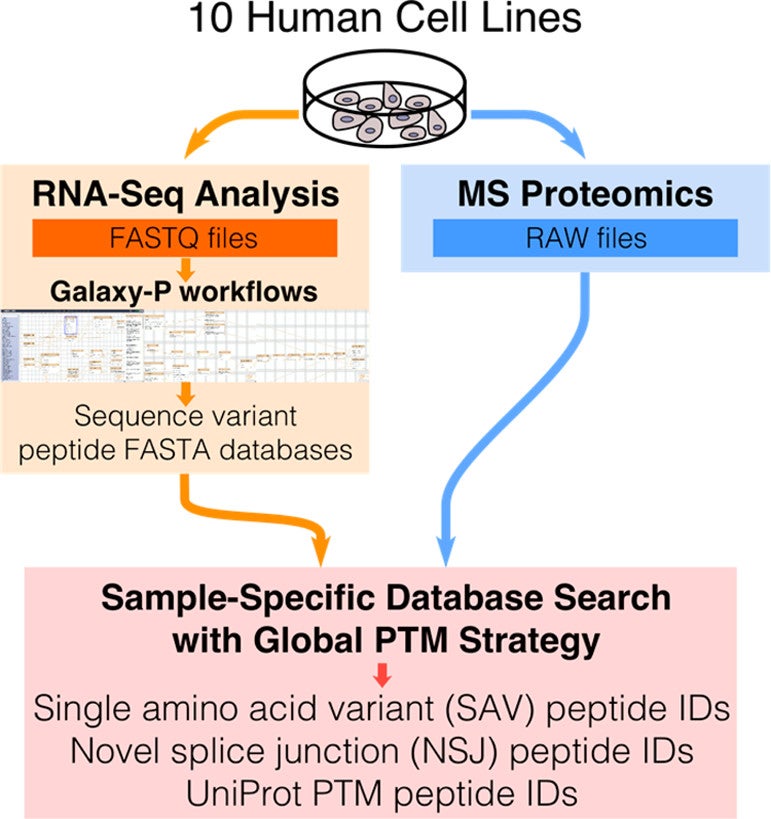
Using Galaxy-P to leverage RNA-Seq for the discovery of novel protein variations
Gloria M Sheynkman, James E Johnson, Pratik D Jagtap, Michael R Shortreed, Getiria Onsongo, Brian L Frey, Timothy J Griffin, & Lloyd M Smith. 2014. BMC Genomics. 15: 703.
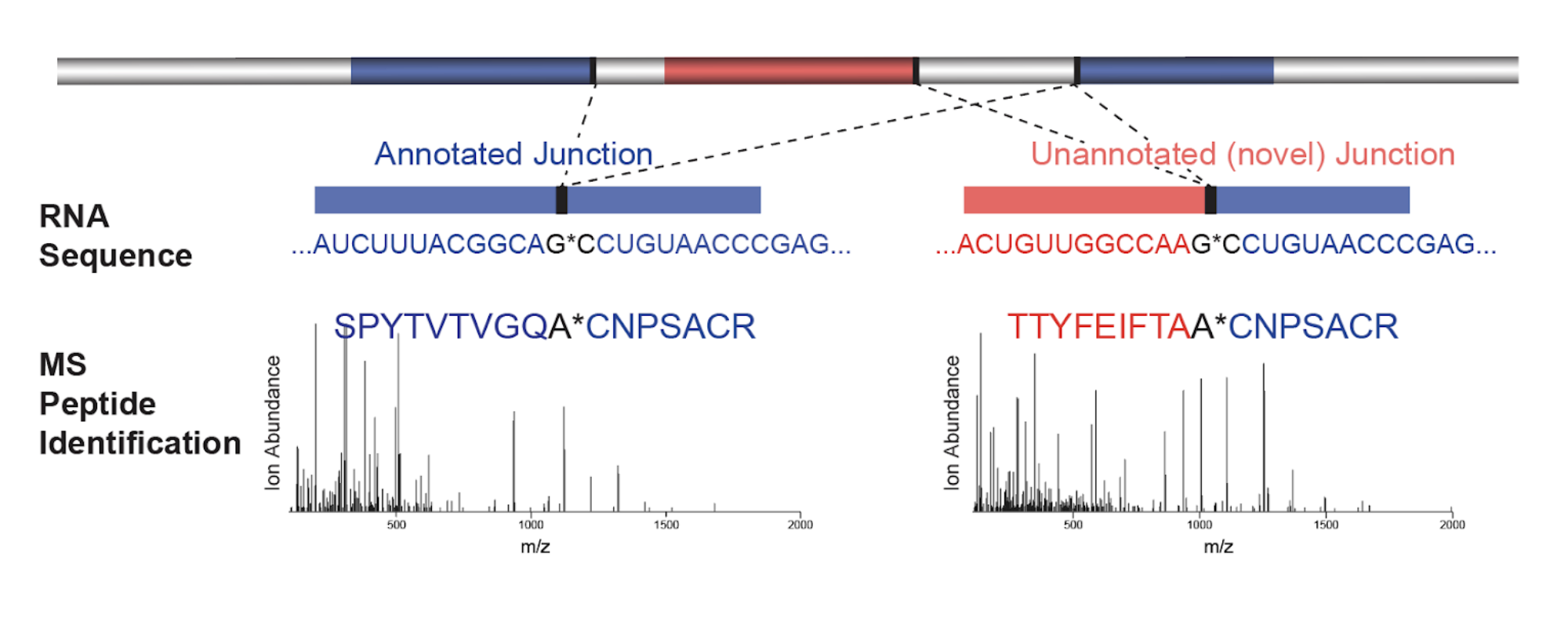
Discovery and mass spectrometric analysis of novel splice-junction peptides using RNA-Seq
Gloria M. Sheynkman, Michael R. Shortreed, Brian L. Frey, Lloyd M. Smith . 2013. Molecular & Cellular Proteomics. 12(8): 2341-53.
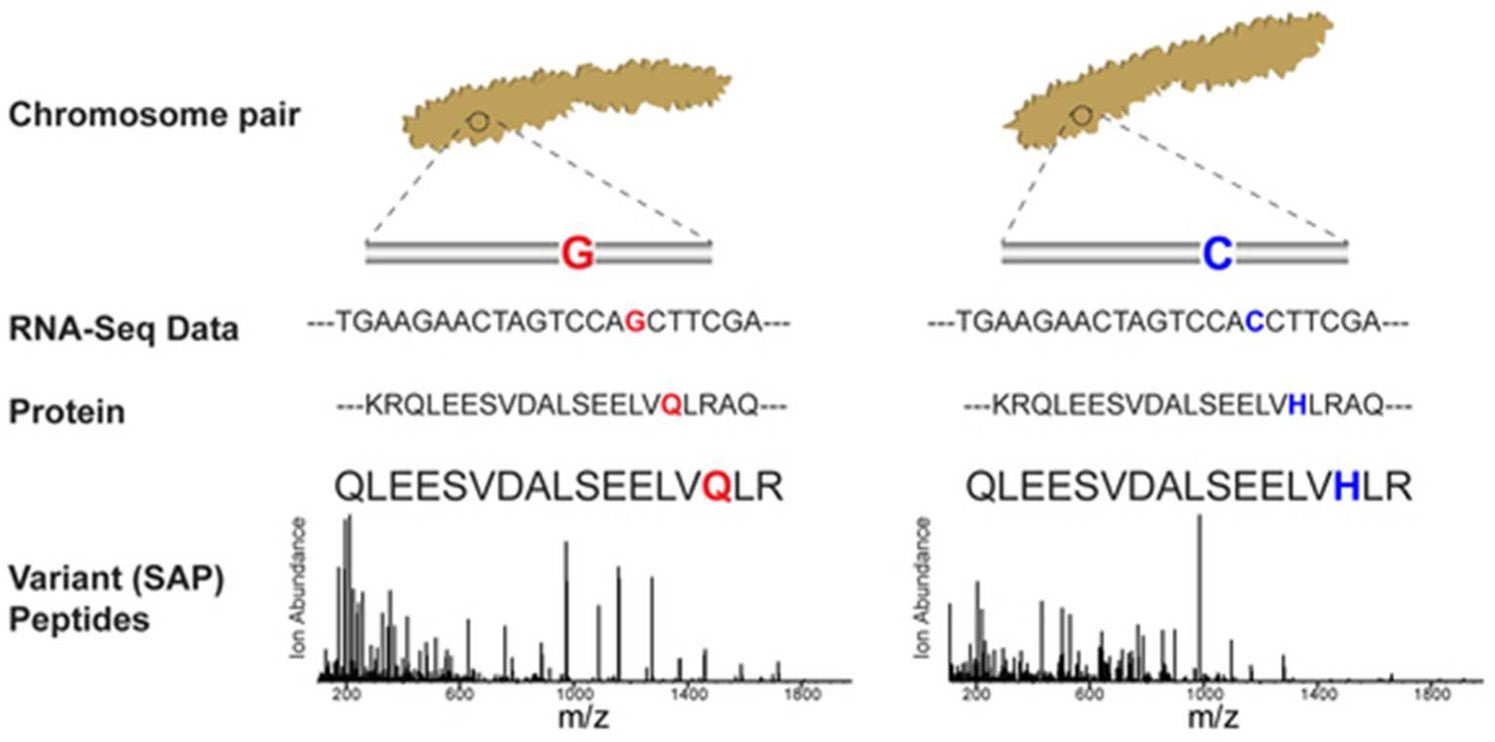
Large-scale mass spectrometric detection of variant peptides resulting from nonsynonymous nucleotide differences
Gloria M. Sheynkman, Michael R. Shortreed, Brian L. Frey, Mark Scalf, Lloyd M. Smith. 2013. Journal of Proteome Research. 13(1): 228-40.
Absolute quantification of prion protein (90-231) using stable isotope-labeled chymotryptic peptide standards in a LC-MRM AQUA workflow
Robert Sturm, Gloria Sheynkman, Clarissa Booth, Lloyd M. Smith, Joel A. Pedersen, & Lingjun Li. 2012. Journal of the American Society for Mass Spectrometry. 23(9): 1522-33.
Sequence-specific capture of protein-DNA complexes for mass spectrometric protein identification
Cheng-Hsien Wu, Siyuan Chen, Michael R. Shortreed, Gloria M. Kreitinger, Yuan Yuan, Brian L. Frey, Yi Zhang, Shama Mirza, Lisa A. Cirillo, Michael Olivier, Lloyd M. Smith. 2011. PloS One. 6(10): e26217.
Alkylating tryptic peptides to enhance electrospray ionization mass spectrometry analysis
Suzanne E. Kulevich, Brian L. Frey, Gloria Kreitinger, Lloyd M. Smith. 2010. Analytical Chemistry. 82(24): 10135-42.
Metabolism of a highly selective gelatinase inhibitor generates active metabolite
Mijoon Lee, Adriel Villegas-Estrada, Giuseppe Celenza, Bill Boggess, Marta Toth, Gloria Kreitinger, Christopher Forbes, Rafael Fridman, Shahriar Mobashery, Mayland Chang. 2007. Chemical Biology & Drug Design. 70(5): 371-82.