Selected Publications
Long-read proteogenomics to connect disease-associated sQTLs to 2 the protein isoform effectors of disease
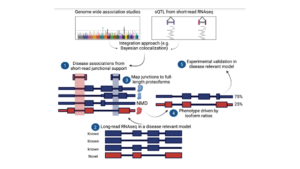
Abdullah Abood, Larry D. Mesner, Erin D. Jeffery , Mayank Murali , Micah Lehe , Jamie Saquing , Charles R. Farber, Gloria M. Sheynkman,
Systematic assessment of long-read RNA-seq methods for transcript identification and quantification
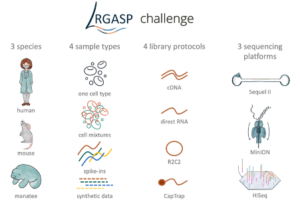
Pardo-Palacios FJ, Wang D, Reese F, Diekhans M, Carbonell-Sala S, Williams B, Loveland JE, De María M, Adams MS, Balderrama-Gutierrez G, Behera AK, Gonzalez JM, Hunt T, Lagarde J, Liang CE, Li H, Jerryd Meade M, Moraga Amador DA, Prjibelski AD, Birol I, Bostan H, Brooks AM, Hasan Çelik M, Chen Y, Du MRM, Felton C, Göke J, Hafezqorani S, Herwig R, Kawaji H, Lee J, Liang Li J, Lienhard M, Mikheenko A, Mulligan D, Ming Nip K, Pertea M, Ritchie ME, Sim AD, Tang AD, Kei Wan Y, Wang C, Wong BY, Yang C, Barnes I, Berry A, Capella S, Dhillon N, Fernandez-Gonzalez JM, Ferrández-Peral L, Garcia-Reyero N, Goetz S, Hernández-Ferrer C, Kondratova L, Liu T, Martinez-Martin A, Menor C, Mestre-Tomás J, Mudge JM, Panayotova NG, Paniagua A, Repchevsky D, Rouchka E, Saint-John B, Sapena E, Sheynkman L, Laird Smith M, Suner MM, Takahashi H, Youngworth IA, Carninci P, Denslow ND, Guigó R, Hunter ME, Tilgner HU, Wold BJ, Vollmers C, Frankish A, Fai Au K, Sheynkman GM, Mortazavi A, Conesa A, Brooks AN. Systematic assessment of long-read RNA-seq methods for transcript identification and quantification. bioRxiv [Preprint]. 2023 Jul 27:2023.07.25.550582. doi: 10.1101/2023.07.25.550582. PMID: 37546854; PMCID: PMC10402094.

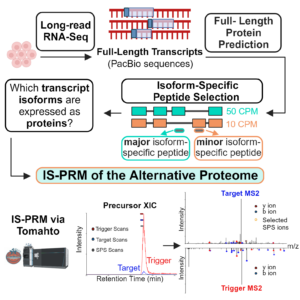
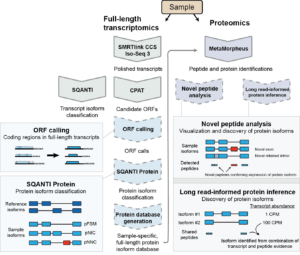
Miller RM, Jordan BT, Mehlferber MM, Jeffery ED, Chatzipantsiou C, Kaur S, Millikin RJ, Dai Y, Tiberi S, Castaldi PJ, Shortreed MR, Luckey CJ, Conesa A, Smith LM, Deslattes Mays A, Sheynkman GM. Enhanced protein isoform characterization through long-read proteogenomics. Genome Biol. 2022 Mar 3;23(1):69. PMID: 35241129
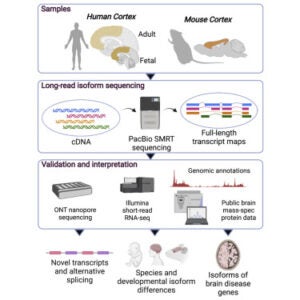
Leung SK, Jeffries AR, Castanho I, Jordan BT, Moore K, Davies JP, Dempster EL, Bray NJ, O’Neill P, Tseng E, Ahmed Z, Collier DA, Jeffery ED, Prabhakar S, Schalkwyk L, Jops C, Gandal MJ, Sheynkman GM, Hannon E, Mill J. Full-length transcript sequencing of human and mouse cerebral cortex identifies widespread isoform diversity and alternative splicing. Cell Rep. 2021 Nov 16;37(7):110022. PMID: 34788620
Saferali A, Xu Z, Sheynkman GM, Hersh CP, Cho MH, Silverman EK, Laederach A, Vollmers C, Castaldi PJ. Characterization of a COPD-Associated NPNT Functional Splicing Genetic Variant in Human Lung Tissue via Long-Read Sequencing. medRxiv [Preprint]. 2020 Nov 3:2020.10.20.20203927. PMID: 33173926
Sheynkman GM, Tuttle KS, Laval F, Tseng E, Underwood JG, Yu L, Dong D, Smith ML, Sebra R, Willems L, Hao T, Calderwood MA, Hill DE, Vidal M, ORF Capture-Seq as a versatile method for targeted identification of full-length isoforms., 2020; Nature communications. 11(1) 2326. PMID: 32393825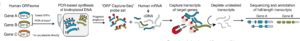
Luck K, Kim DK, Lambourne L, Spirohn K, Begg BE, Bian W, Brignall R, Cafarelli T, Campos-Laborie FJ, Charloteaux B, Choi D, Coté AG, Daley M, Deimling S, Desbuleux A, Dricot A, Gebbia M, Hardy MF, Kishore N, Knapp JJ, Kovács IA, Lemmens I, Mee MW, Mellor JC, Pollis C, Pons C, Richardson AD, Schlabach S, Teeking B, Yadav A, Babor M, Balcha D, Basha O, Bowman-Colin C, Chin SF, Choi SG, Colabella C, Coppin G, D’Amata C, De Ridder D, De Rouck S, Duran-Frigola M, Ennajdaoui H, Goebels F, Goehring L, Gopal A, Haddad G, Hatchi E, Helmy M, Jacob Y, Kassa Y, Landini S, Li R, van Lieshout N, MacWilliams A, Markey D, Paulson JN, Rangarajan S, Rasla J, Rayhan A, Rolland T, San-Miguel A, Shen Y, Sheykhkarimli D, Sheynkman GM, Simonovsky E, Taşan M, Tejeda A, Tropepe V, Twizere JC, Wang Y, Weatheritt RJ, Weile J, Xia Y, Yang X, Yeger-Lotem E, Zhong Q, Aloy P, Bader GD, De Las Rivas J, Gaudet S, Hao T, Rak J, Tavernier J, Hill DE, Vidal M, Roth FP, Calderwood MA. A reference map of the human binary protein interactome., 2020; Nature. 580(7803) 402-408. PMID: 32296183
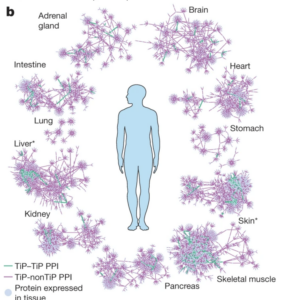
Luck K, Sheynkman GM, Zhang I, Vidal M, Proteome-Scale Human Interactomics., 2017; Trends in biochemical sciences. 42(5) 342-354. PMID: 28284537
Sheynkman GM, Shortreed MR, Cesnik AJ, Smith LM, Proteogenomics: Integrating Next-Generation Sequencing and Mass Spectrometry to Characterize Human Proteomic Variation., 2016; Annual review of analytical chemistry (Palo Alto, Calif.). 9(1) 521-45. PMID: 27049631
Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray RR, Spirohn K, Begg BE, Duran-Frigola M, MacWilliams A, Pevzner SJ, Zhong Q, Trigg SA, Tam S, Ghamsari L, Sahni N, Yi S, Rodriguez MD, Balcha D, Tan G, Costanzo M, Andrews B, Boone C, Zhou XJ, Salehi-Ashtiani K, Charloteaux B, Chen AA, Calderwood MA, Aloy P, Roth FP, Hill DE, Iakoucheva LM, Xia Y, Vidal M, Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing., 2016; Cell. 164(4) 805-17. PMID: 26871637
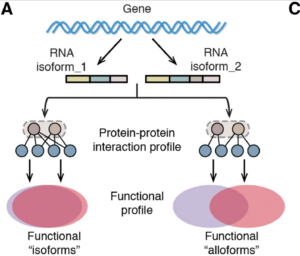
Sheynkman GM, Shortreed MR, Frey BL, Smith LM, Discovery and mass spectrometric analysis of novel splice-junction peptides using RNA-Seq., 2013; Molecular & cellular proteomics : MCP. 12(8) 2341-53. PMID: 23629695
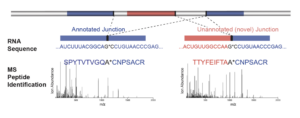
Sheynkman GM, Shortreed MR, Frey BL, Scalf M, Smith LM, Large-scale mass spectrometric detection of variant peptides resulting from nonsynonymous nucleotide differences., 2013; Journal of proteome research. 13(1) 228-40. PMID: 24175627