Vande Pol Lab Research Opportunities
Scott Vande Pol, M.D., Ph.D., D (ABMM)
 Associate Professor of Pathology
Associate Professor of Pathology
vandepol@virginia.edu
Laboratory Interests:
Papillomaviruses are the most prevalent sexually transmitted disease and the leading infectious cause of cancer in the US, with a yearly economic impact of about $2.9 billion (2). Despite a prophylactic vaccine, lethal clinical disease will be a significant problem for many decades to come world-wide. Alpha genus human papillomaviruses (HPVs) cause benign squamous epitheliomas in which the virus persists and replicates. With HPV types termed “high-risk”, the initially benign papilloma may evolve over time and develop into a malignancy. “Low-risk” HPV types very rarely progress to malignancy, but can cause serious, and even life-threatening medical complications due to the size, numbers, and locations of the benign papillomas.
I study the structure and biology of papillomavirus oncoproteins. We study papillomavirus oncoproteins from both human and animals to uncover their biological logic and the mechanism by which they transform cells and promote cancer. By comparing oncoproteins from diverse papillomaviruses, we have begun to understand how these oncoproteins physically interact with their cellular targets, and influence host cell signaling pathways.
Structure-Function Analysis of E6 Proteins.
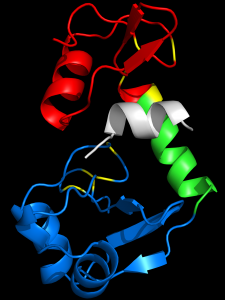 Our studies of the bovine papillomavirus E6 oncoprotein were the first to identify the mechanism by which E6 associates with cellular proteins: by docking on an alpha-helical LXXLL peptide motif in the target cellular protein. We were able to show by intracellular competitive binding experiments that the E6-LXXLL interaction was essential for biological activity of E6 proteins. We showed that the E6-LXXLL interaction solubilized E6, rendering the protein suitable for structural studies. The association of HPV-16 E6 with LXXLL peptides causes a conformational change in E6 that confers E6 solubility and stability. For the prior 30 years, academic and industrial labs had failed to obtain a structure of E6 because in the absence of a bound LXXLL peptide, E6 is insoluble. We used this to recruit collaborators in Strasbourg France (Dr Gilles Trave and associates) to produce and crystalize bovine and human papillomavirus E6 bound to LXXLL peptides. We were further able to show that when E6 binds its LXXLL peptide, it adopts a conformation that allows direct interaction with the p53 tumor suppressor.
Our studies of the bovine papillomavirus E6 oncoprotein were the first to identify the mechanism by which E6 associates with cellular proteins: by docking on an alpha-helical LXXLL peptide motif in the target cellular protein. We were able to show by intracellular competitive binding experiments that the E6-LXXLL interaction was essential for biological activity of E6 proteins. We showed that the E6-LXXLL interaction solubilized E6, rendering the protein suitable for structural studies. The association of HPV-16 E6 with LXXLL peptides causes a conformational change in E6 that confers E6 solubility and stability. For the prior 30 years, academic and industrial labs had failed to obtain a structure of E6 because in the absence of a bound LXXLL peptide, E6 is insoluble. We used this to recruit collaborators in Strasbourg France (Dr Gilles Trave and associates) to produce and crystalize bovine and human papillomavirus E6 bound to LXXLL peptides. We were further able to show that when E6 binds its LXXLL peptide, it adopts a conformation that allows direct interaction with the p53 tumor suppressor.
Primary Cellular Targets of E6 Proteins, and Evolutionary Organization of Papillomaviruses
While high risk HPV E6 oncoproteins are best known for binding the cellular ubiquitin ligase E6AP and then targeting p53 for degradation, most other human and animal papillomavirus E6 proteins do not bind E6AP. So what then is the target of E6 proteins that do not interact with E6AP? Our studies first identified (followed closely by the Munger and Howley labs) the most frequent cellular target of diverse human and animal papillomaviruses E6 oncoproteins: the MAML family of transcriptional co-activators that mediate Notch signaling. In squamous epithelia, Notch signaling acts as a tumor suppressor. E6 associates with MAML1 through docking on a LXXLL peptide in MAML1 and down-regulates Notch signaling. We have evidence that this interaction between E6 and MAML1 is the predominant interaction of papillomavirus E6 oncoproteins in cutaneous papillomavirus infections of humans and diverse species of animals.
Secondary cellular targets of E6AP-associated E6 proteins
As noted above, E6 proteins that interact with E6AP first bind to the LXXLL motif of E6AP, then change conformation to interact with a secondary target such as p53. We have used proteomic approaches to identify additional secondary cellular proteins that are targeted for degradation by E6 plus E6AP. We were the first to identify tyrosine phosphatases as targets of papillomavirus oncoproteins. One of these targets, PTPN3, confers decreased growth-factor dependence of keratinocytes when targeted by E6. We are expanding the known E6 interactions with cellular PDZ proteins and recently showed that E6 interaction with cellular PDZ proteins regulates the Hippo pathway effector YAP1. Of exceptional interest, we have recently identified a cellular protein that regulates WNT signaling as the first described cellular protein that is targeted for degradation by both high-risk, low-risk, and diverse animal E6 proteins, SLC9A3R1; this work is currently under preparation for publication. This is the first cellular protein described to be a target of diverse genera of E6 proteins.
Rotation Projects
Rotations will be focused upon the mechanism by which E6 proteins trigger ubiquitin ligase activity of the cellular E6AP ubiquitin ligase. An alternative lab rotation would address some aspect of the interaction of E6 proteins with the cellular adapter protein NHERF1.
What general knowledge and techniques would you would learn during a rotation
Students will acquire knowledge in viral oncogenesis and papillomaviruses. Techniques that would likely be utilized would include the preparation of lentiviruses, tissue culture expression analysis, molecular cloning, yeast genetics, and western blotting.
References
- Zanier K, Charbonnier S, Sidi AO, McEwen AG, Ferrario MG, Poussin-Courmontagne P, Cura V, Brimer N, Babah KO, Ansari T, Muller I, Stote RH, Cavarelli J, Vande Pol* S, Travé* G. Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science. 2013 Feb 8;339(6120):694-8. PMCID: PMC3899395. *corresponding authors.
- Brimer, N., and S.B.Vande Pol. Papillomavirus E6 PDZ interactions can be replaced by repression of p53 to promote episomal HPV genome maintenance. J. Virol. Mar;88(5):3027-30. doi: 10.1128/JVI.02360-13. Epub 2013 Dec 18.. PMC3958089 PMID: 24352452.
- Vande Pol, S.B. “Papillomavirus E6 oncoproteins take common structural approaches to solve different biological problems. PLoS-Pathogens. 2015 Oct 15;11(10):e1005138. doi: 10.1371/journal.ppat.1005138. eCollection 2015 PMC4607424
- Martinez-Zapien D, Ruiz FX, Poirson J, Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Vande Pol S, Podjarny A, Travé G, Zanier K. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016 Jan 28;529(7587):541-5. doi: 10.1038/nature16481. Epub 2016 Jan 20. PMID: 26789255; PMCID: PMC4853763.
- Petropoulos C1, Oddou C1, Emadali A2, Hiriart-Bryant E1, Boyault C1, Faurobert E1, Vande Pol S3, Kim-Kaneyama JR4, Kraut A5, Coute Y5, Block M1, Albiges-Rizo C1, Destaing O6. Roles of paxillin family members in adhesion and ECM degradation coupling at invadosomes. J Cell Biol. 2016 Jun 6;213(5):585-99. doi: 10.1083/jcb.201510036. PMCID: PMC4896053
- Strickland, S., and Vande Pol, S.B. The Human Papillomavirus Type 16 E7 Oncoprotein Attenuates AKT Signaling to Promote IRES Dependent Translation and expression of c-MYC. J. Virol. 2016 May 27;9(12):5611-21. PMCID: 27030265
- Nicole Brimer1, Camille M. Drews1, and Scott B. Vande Pol. Association of papillomavirus E6 proteins with either MAML1 or E6AP clusters E6 proteins by structure, function, and evolutionary relatedness. PLoS Pathogens, accepted 12_19_17. PMCID: PMC5760104
- Sydney Strickland1, Nicole Brimer, Charles Lyons1*, and Scott B. Vande Pol1. Human Papillomavirus E6 interaction with cellular PDZ domain proteins modulates YAP nuclear localization. 2018 Virology. Mar;516:127-138. doi: 10.1016/j.virol.2018.01.003. Epub 2018 Jan 12. PMCID: PMC5826896.
- Camille Drews, Samuel Case, and Scott Vande Pol. “E6 proteins from high-risk HPV, low-risk HPV, and animal papillomaviruses activate the Wnt/β-catenin pathway through E6AP-dependent degradation of NHERF1″. PLoS Pathogens, Published online 2019 Apr 19. PMCID: PMC6493770
- Camille Drews, Nicole Brimer, and Scott Vande Pol. ““Multiple regions of E6AP (UBE3A) contribute to interaction with papillomavirus E6 proteins and the activation of ubiquitin ligase activity.” January 2020 PLoS Pathogens 16(1):e1008295. PMCID: PMC6999913
