Novel Mass-Spectrometry Technologies for Proteoform Detection
Novel Mass-Spectrometry Technologies for Proteoform Detection

The challenge of proteoform detection
A proteoform is variation of a protein sequence, whether that be caused by alternative splicing, single nucleotide polymorphism (or other genetic mutation), or post-translational modification. The challenge in distinguishing proteoforms from biological samples is that different proteoforms often share the vast majority of amino acid sequence. In the Sheynkman lab, we use bioinformatic tools to select the distinguishing regions of amino acids among proteoforms to use as target peptides for confirming their presence in samples.
Peptides that are specific to alternative isoforms can also be overlooked due to lower proteotypicity (or amenability to MS instrumentation) or lack of representation in publicly available search databases. We can circumvent these issues using advanced targeted MS methods that allow us to target disease-relevant isoform-specific peptides that are predicted directly from long-read RNA sequencing data.
MS Instrument methods for targeting alternative-splicing (AS) derived isoforms
While traditional mass spectrometry methods like Data-Dependent Acquisition (DDA) are powerful tools in proteomics, they tend to favor the detection of higher-abundance ions, making them less suited for the identification of alternative protein isoforms, which are often present at lower concentrations. Targeting methods have better sensitivity and can be used when there is a priori knowledge of the peptides of interest. Parallel reaction monitoring (PRM) is often used as a targeting method in proteomics.
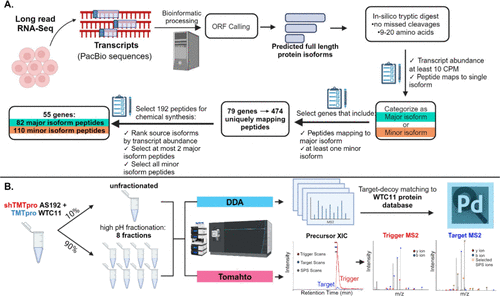
IS-PRM to assay hundreds of targets
To further increase the scale of how many isoform-distinguishing peptides we can target in one assay, one of the most powerful methods at our disposal is internal standard parallel reaction monitoring or IS-PRM. This method sensitively initiates MS2 collection of target peptides based on detection of synthetic trigger peptides that match the amino acid sequence of targets but are distinguishable by a mass offset. This allows us to assay hundreds of isoform-specific peptides in one MS run and vastly outperform DDA at alternative protein isoform detection.
Here is a recent publication from our lab that demonstrates the use of IS-PRM to identify AS isoforms from WTC11 cells.
IS-PRM-based peptide targeting informed by long-read sequencing for alternative proteome detection
Jennifer A. Korchak, Erin D. Jeffery, Saikat Bandyopadhyay, Ben T. Jordan, Micah Lehe, Emily F. Watts, Aidan Fenix, Mathias Wilhelm, Gloria M. Sheynkman
