Human Disease Genetics
 Human disease genetics is the study of how genetic variations contribute to diseases and health conditions in humans. This field encompasses a range of topics, including the identification of genes associated with specific diseases, the understanding of how these genes interact with environmental factors, and the mechanisms by which genetic mutations lead to disease. Some key areas that our lab’s research focuses on are splicing quantitative trait loci (sQTL), expression quantitative trait loci (eQTL), and other forms of genetic variance.
Human disease genetics is the study of how genetic variations contribute to diseases and health conditions in humans. This field encompasses a range of topics, including the identification of genes associated with specific diseases, the understanding of how these genes interact with environmental factors, and the mechanisms by which genetic mutations lead to disease. Some key areas that our lab’s research focuses on are splicing quantitative trait loci (sQTL), expression quantitative trait loci (eQTL), and other forms of genetic variance.
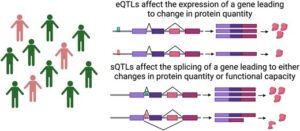
An sQTL is a genetic variant that is associated with changes in the splicing ratios of transcripts. They can be caused by single nucleotide polymorphisms (SNPs) that alter splicing patterns or alternative splicing (AS), where exons from the same gene are joined in different combinations. As sQTLs lead to the production of different isoforms, and these isoforms can have varying functions, they may contribute to disease processes in conditions.
An eQTL is a chromosomal region where genetic variants are associated with the expression levels of specific genes. eQTLs influence the expression levels of genes, upregulating or downregulating specific genes, leading to varying levels of isoforms that may lead to disease.
Genetic variants, also known as mutations, are major determinants of susceptibility to disease and response to treatment, and major advances in sequencing technologies has enabled the identification genetic variants in each genome.
We use a wide variety of bioinformatic approaches to identify the genetic mechanisms of disease in a variety of disease communities, including cancer, osteoporosis (BMD), chronic obstructive pulmonary disease (COPD), and coronary artery disease (CAD).
Here are some recent examples of our lab’s related research (Sheynkman Lab members are underlined):
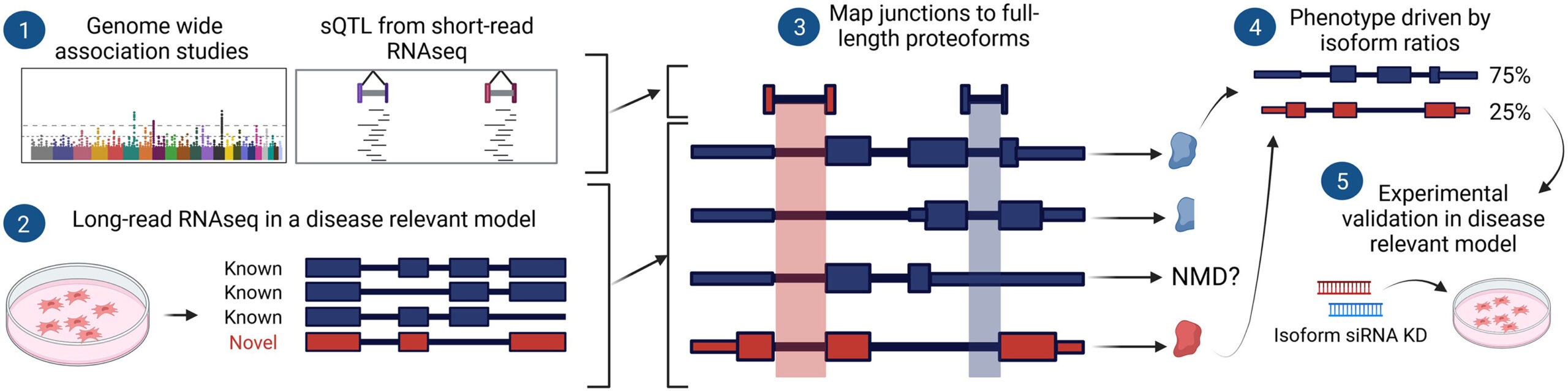
Abdullah Abood, Larry D. Mesner, Erin D. Jeffery , Mayank Murali , Micah Lehe , Jamie Saquing , Charles R. Farber, Gloria M. Sheynkman. 2024. Long-read proteogenomics to connect disease-associated sQTLs to the protein isoform effectos of disease

