Bekiranov Lab
Machine Learning/AI for Biomedical Applications
Development of innovative advanced machine learning approaches including knowledge-guided meta-learning explainable machine learning frameworks that take advantage of the relationships between genes, cell types, clinical variables, cardiovascular disease (CVD), cancers, etc. to build models that can predict clinical outcomes (e.g., CVD or cancer patient survival using multi-omics data as input) or functional genomic outputs (e.g., gene expression levels using regulatory factor data as input). I am developing these approaches independently as well as in collaboration with Dr. Aidong Zhang in UVA’s Computer Science department. My lab has developed and applied machine learning approaches—non-negative matrix factorization, regression (e.g., MARS, multi-linear, lasso), unsupervised clustering, etc.—using epigenomic data (i.e., genome-wide profiles of histone modifications, histone variants, DNA methylation, DNAse hypersensitive sites, nucleosome occupancy and transcription factor binding) to elucidate how epigenomic factors control processes on the DNA template including transcription and DNA replication. With, Dr. Coleen McNamara, we have also recently developed, tested and compared deep learning and ensemble tree models that predict risk of cardiovascular disease (CVD) using standard clinical variables and a CVD SNP as input. I am currently the Director of Bioinformatics of the Precision Immunomedicine (iPRIME) initiative whose broad goals are to develop precision medicine approaches and predict risk of various CVD endpoints by generating and performing AI/ML and integrative analysis of single-cell immune cell signature (derived from mass cytometry), genome-wide SNP, advanced imaging and clinical data.
Mathematical Modeling of Transcription Factor-Chromatin Binding Dynamics
Development of deterministic and stochastic mathematical models of transcription factor binding to chromatin and nucleosome turnover using data from assays which measure the dynamics of these events in vivo and enable extraction of kinetic parameters. These assays include crosslinking kinetic (CLK), which Dr. David Auble and I developed, Competition ChIP, Anchor Away and Auxin-Inducible Degron Systems. The long-term goals of this project are to develop stochastic mathematical models of pre-initiation complex (PIC) formation, determine the epigenetic and genetic factors that regulate PIC dynamics and integrate these models with stochastic models of transcription whose dynamics have been characterized by highly stochastic, infrequent bursts from imaging and scRNA-seq data.
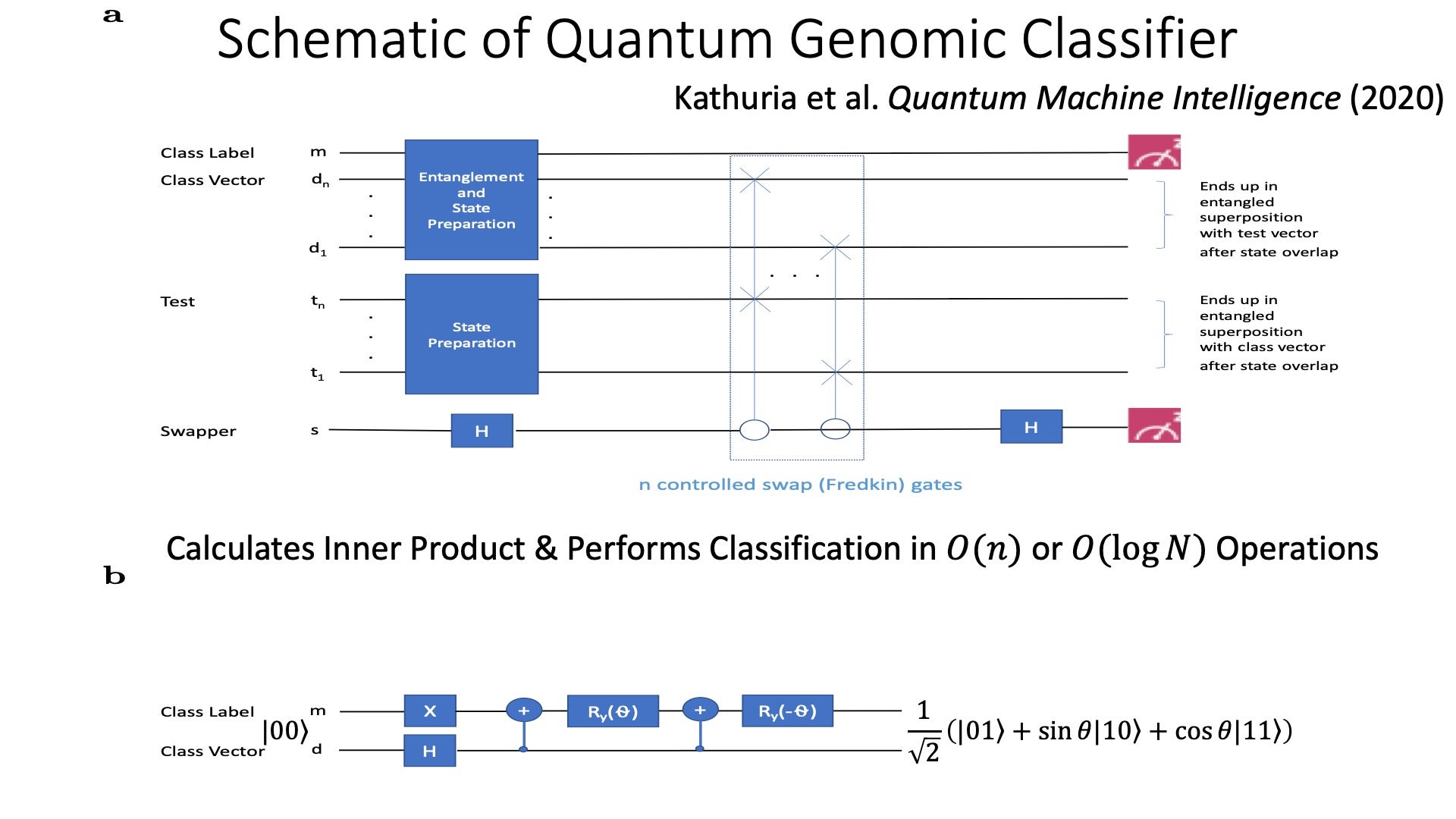
Quantum Computing and Quantum Machine Learning
Development and implementation of quantum machine learning algorithms for genomic applications. My lab recently developed an inner product-based quantum classifier which we show is exponentially more efficient compared to its classical counterpart and implement on 5-qubit and 14-qubit IBM quantum computers. In the short term, we are extending this work to building and implementing kernel-based quantum classifiers with more complex decision boundaries as well as addressing the data input problem (e.g., by encoding data in phases of complex probability amplitudes). Our long-term goals are to develop and implement quantum machine learning algorithms that demonstrate quantum advantage. Notably, this work has gained national and international interest in that developing quantum computing algorithms for the biomedical/genomics domain is currently extremely rare.
