Equipment
Instrument List
The Chromium instruments performs the integral first step in constructing 10x Genomics single cell/nuclei NGS libraries by creating the emulsion bubbles in which single cells are partitioned and individually tagged with a unique barcode for downstream analysis. The microfluidic chips used for the emulsion can hold multiple samples simultaneously.
The core has upgraded its single cell service instrument from the 10X Chromium Controller to the 10X Chromium X. The chromium X can partition hundreds of thousands of fresh, frozen, or fixed cells in a single run. The core’s instrument is routinely used for projects ranging from small pilot studies to the highest-throughput analysis, delivering readouts for gene expression, chromatin accessibility, cell surface proteins, immune clonotype, antigen specificity, and CRISPR perturbation screens.

Thanks to generous support from the UVA Cancer center, a second NextSeq2000 is now available in the GATC. The NextSeq 2000 are Illumina Next Generation Sequencing (NGS) benchtop instrumentations. They operates the P1, P2 and P3 patterned flow cell which have output of 120, 400, and 1,000 million reads per run. In addition to running the standard Illumina sequencing by synthesis chemistry, the two core’s NextSeq 2000 have been recently upgraded to run the new XLEAP-SBS chemistry which will enable faster, higher quality, and more robust runs. XLEAP-SBS nucleotides use state-of-the-art dyes and novel linkers and blocks that are more resistant to heat, show a 50× reduction in hydrolysis, and 2.5× faster block cleavage to reduce phasing and prephasing. At this time, the XLEAP-SBS chemistry is available with the P4 flow cell with output of 2 Billion reads per run. XLEAP-SBS chemistry on P1, P2, P3 will be available Q2 2024.
More information can be found on the Planning an NGS Experiment page.

Applications
- Whole-genome resequencing
- Targeted resequencing, including exome panels and custom enrichment panels (100s kb–Mbs)
- Whole-genome de novo sequencing
- Sequencing of bisulfite-treated DNA (Methyl-Seq)
- RNA sequencing (RNA-Seq), including mRNA sequencing and total RNA sequencing (coding and noncoding)
- Genotyping by sequencing
- Nucleosome positioning and chromatin structure studies
- ChIP-Seq: studying sequence-specific protein–RNA interactions
- Sequencing of ancient DNA samples
- Single-cell applications including RNA-Seq, VDJ, Cell surface expression, ATAC-seq
- Linked long-read applications
[/one_half]
The miSeq desktop sequencer allows access to more focused applications such as microbiome sequencing (bacterial and fungal), amplicon sequencing, metagenomics, and small genome sequencing, starting with 10ng of DNA. New MiSeq reagents enable up to 25 million sequencing reads and 2×300 bp read lengths.


The QIAcuity is a nanoplate-based system which seamlessly integrates a standard dPCR workflow of partitioning, thermocycling and imaging into a walk-away automated platform with minimal hands-on time.The QIAcuity system makes absolute quantification accessible, affordable and easy for all laboratories. The walk-away automation integrates and streamlines the entire digital PCR workflow into single instrument with minimal hands-on time. It is also easy to adapt your current qPCR assays to the QIAcuity Digital PCR System. No change in plate handling is required when coming from qPCR, assuring fast assay setup and quick results in under 2 hours.
The QIAcuity Digital PCR System will deliver precise and multiplexed quantification results for all molecular applications including but not limited to the following research areas:
- -Rare mutation detection
- -Copy number variation analysis
- -Gene expression analysis
- -NGS library quantification
- -Pathogen detection
- -Genotyping
- -miRNA research

The QuantStudio 6 performs both SYBR Green and TaqMan qPCR. Real-time quantitative PCR (qPCR) is a highly sensitive and specific method for SNP genotyping and for amplifying, detecting and quantitating target transcripts. The QuantStudio 6-Flex enables detection as small as 1.5-fold changes in singleplex reactions and with 10 logs dynamic range. It can also be used to identify new variants quickly and accurately with high resolution melt.
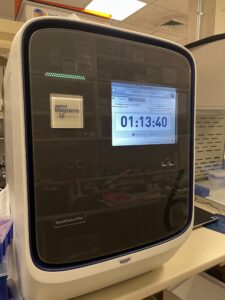
The Agilent 4200 TapeStation system is an automated electrophoresis tool for assessing size, quality, and concentration of up to 96 DNA or RNA samples per run. This instrument is commonly used to QC RNA prior to RNAseq library prep and to QC finished NGS libraries. The core currently stocks a variety of kits for analyzing RNA and DNA of both high and low concentrations. This instrument is both Core and user operated, but requires user training before use.
The Agilent Bioanalyzer uses a microfluidic chip to perform similar analyses to the TapeStation. This instrument is currently only available for special request runs of a type that are not supported by the Agilent TapeStation. Please inquire.




